Plasmonic nanowire interstice sensor for the diagnosis of prostate cancer
Extracellular microRNAs recently provided valuable information including the site and the status of cancers. miR141 and miR375 are the most pronounced biomarkers for the diagnosis of high-risk prostate cancer. Here, we describe attomolar detection of miR141 and miR375 released from living prostate cancer cells through the use of a plasmonic nanowire interstice (PNI) sensor.
by Dr Taejoon Kang and Professor Bongsoo Kim
Background
Prostate-specific antigen
Prostate cancer (PC) represents 27% of all cancers in men and the second leading cause of cancer death for men worldwide [1]. In 2017 for the USA alone, there were approximately 161 360 cases of PC. PC has been diagnosed by digital rectal examination and the prostate-specific antigen (PSA) test. PSA is the only tissue-specific biomarker that can aid the early diagnosis of PC. The PSA blood test, however, has limited diagnostic accuracy for PC because PSA can be increased owing to other factors including benign prostatic hyperplasia or prostatitis as well as PC. The US Preventive Services Task Force even recommended that physicians should not routinely perform PC screening based on serum PSA levels [2]. Clearly, new biomarkers are needed to overcome this problem.
Recently, it has been reported that the level of free PSA (f-PSA) is decreased in men who have PC compared with those with benign conditions [3]. Therefore, various immunoassay technologies including enzyme-linked immunosorbent assay, fluorescence immunoassay, surface plasmon resonance (SPR), electrochemical immunosensor, dark-field microscopy, chemiluminescence, surface-enhanced Raman scattering (SERS), and dynamic light scattering have been employed for the quantitative analysis of f-PSA [3].
RNAs as prostate cancer biomarkers
Long noncoding RNAs (lncRNAs, ≥200 nucleotides) are often expressed in a disease-, tissue- or developmental-specific manner. Since lncRNAs are highly dysregulated in several cancer types and exhibit a high degree of tissue- and disease-specificity, lncRNAs are regarded as candidates for cancer diagnostic biomarkers [4]. Prostate Cancer Antigen 3 (PCA3) is a prostate-specific lncRNA that is overexpressed by 60- to 100-fold in >90% of prostate tumours compared to benign prostatic tissue. Urinary PCA3 has been used as a diagnostic biomarker for PC with a sensitivity of 58–82% and a specificity of 56–76%. The sensitivity and accuracy of PCA3 are increased when used in combination with α-methylacyl-CoA racemase. Urinary PCA3 is now widely used for PC diagnosis and has been approved by the US Food and Drug Administration (FDA). MicroRNAs (miRNAs) are single-stranded, small, and noncoding RNAs. The expression patterns of miRNAs in tissue and blood samples of patients are often closely associated with disease types and also disease stages, hinting that certain miRNAs can be compelling diagnostic markers [5]. In 2008, it was first reported that the level of miR141 is upregulated in the serum of metastatic PC compared with healthy controls and benign prostatic hyperplasia patients. Since then, miR141 and miR375 have been the most pronounced biomarkers for high-risk PC, including castrate-resistant PC and metastatic PC, which account for approximately 15% of PC diagnoses and have the potential to progress to a lethal phenotype [6].
Detection methods for nucleic acid biomarkers
For the detection of nucleic acid biomarkers, polymerase chain reaction (PCR) is the most extensively used analytical tool. Although PCR is considered the gold standard for the detection of gene biomarkers, it has drawbacks including a long amplification time and the risk of erroneously amplifying contaminants or unrelated gene sequences. To overcome these limitations, PCR-free assays have been developed by taking various sensing approaches such as fluorescence resonance energy transfer, colorimetry, SPR, electrochemistry, SERS, and so on. These methods have contributed to the advance of cancer diagnosis by reducing the drawbacks of PCR. SERS is a fascinating phenomenon that significantly increases the Raman signal of molecules located within nanoscale metallic interstices (hot spots). SERS has been employed for the sensitive detection of nucleic acid because of its single-molecule sensitivity, molecular specificity, and insensitivity to quenching. It is known that the SERS enhancement strongly depends on the detailed morphology of the metal nanostructure. Although a number of promising nanostructures that can be used as efficient SERS-active platforms have been proposed, it still remains a challenging task to develop a practical SERS sensor that can detect multiple nucleic acid biomarkers simultaneously while retaining high sensitivities. The use of single-crystalline noble metal nanowires (NWs) is highly advantageous for SERS-based detection because of their well-defined geometries, atomically smooth surfaces, and simple fabrication process [7]. Previously, we developed several noble metal NW-based SERS sensors including plasmonic nanowire interstice (PNI) sensor, particle-on-NW sensor, NW on a graphene sensor, and nanogap-rich Au NW sensor [8–15]. Among them, PNI nanostructures have been widely employed for the detection of several biochemical molecules. Particularly, by combining the PNI nanostructure with the bi-temperature hybridization process, we were able to detect miRNAs with near-perfect accuracy of single nucleotide polymorphism (SNPs) and at the extremely low detection limit of 100 aM. Here, we introduce a PNI sensor which can detect the extracellular miR141 and miR375 released from living PC cells into a culture medium. This sensor shows an extremely low detection limit of 100 aM for both miR141 and miR375, and a wide dynamic range from 100 aM to 100 pM, covering the typical concentration range of extracellular miRNAs in the bloodstreams of patients. Additionally, the PNI sensor can completely discriminate the single-base mismatches of miR141 and miR375. This excellent sensing capability of the PNI sensor enables the simultaneous detection of miR141 and miR375 released from the cells of PC cell lines (LNCaP and PC-3), showing the potential applicability to a novel PC diagnostic method.
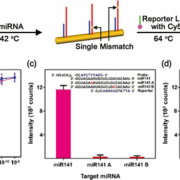
Specific and sensitive detection of miRNA
To accurately determine the expression patterns of miRNAs in biological fluid samples, it is necessary to overcome the inconsistent measurement results caused by low specificities and complicated sensing procedures. For the ultra-specific and ultra-sensitive detection of miRNAs, we applied miRNA-specific bi-temperature hybridizations to Au NW surfaces, where short miRNAs can readily crawl into the narrow hot spots of the PNI sensor for effective SERS detection. The probe locked nucleic acid (LNA)-modified PNI sensors were incubated with miRNAs at 42 °C and subsequently incubated with Cy5-labeled reporter LNA at 64 °C (Fig. 1a). If the target miRNAs have perfectly complementary sequences to both probe and reporter LNAs, sandwiched complexes of probe LNA-miRNA-reporter LNA can be stably formed on a PNI sensor, providing strong SERS signals of Cy5. In contrast, when the sample only contains single-base mismatched miRNAs, little signal was observed. Figure 1(b) displays the intensity of the Cy5 1580 cm−1 band plotted as a function of the miR141 (magenta) and miR375 (blue) concentrations. Both intensities were quite linearly increased throughout the concentration range from 100 aM to 100 pM in spite of the different sequences of miR141 and miR375. To investigate the specificity of a PNI sensor, we prepared four kinds of single-base mismatched miRNAs (miR141 A, miR141 B, miR375 A, and miR375 B). The miR141 A and miR375 A had a mismatched single base on the probe LNA recognition site, respectively, and the miR141 B and miR375 B had a mismatched single base on the reporter LNA recognition site. Figure 1(c,d) shows the plot of Cy5 1580 cm−1 band intensity obtained from the PNI sensors for perfectly matched and single-base mismatched miRNAs. The concentration of all miRNAs was 100 pM. When the single-base mismatched miRNAs (miR141 A, B and miR375 A, B) were present, featureless SERS signals were obtained from the PNI sensors. In contrast, significantly strong SERS signals were measured from the PNI sensors in the presence of miR141 and miR375 with intact sequences. In the miRNA sensing procedure using the PNI sensor, the unstable single-base mismatched miRNA–LNA hybridized structures were destroyed at the temperature over Tm. Therefore, we near-perfectly excluded the possibility of detecting single-base mismatched miRNAs.
Detection of miRNAs released from cells in culture
The PNI sensors were also employed to detect miR141 and miR375 released from the living PC cells. We prepared four types of media in which different human cancer cell lines were cultured. The cultured cell lines were LNCaP (PC cells), PC-3 (PC cells), RWPE-1 (noncancerous prostate epithelial cells), and HeLa (cervical cancer cells). For the detection of miR141 and miR375 using PNI sensors, the total extracellular miRNA released from the cells into the media were isolated and purified. Figure 2(a,b) represent the extracellular miR141 and miR375 levels determined by the PNI sensor for LNCaP, PC-3, RWPE-1, and HeLa, respectively. The levels of miR141 and miR375 in LNCaP and PC-3 culture supernatants were higher than those in RWPE-1 and HeLa, indicating that the PNI sensor can detect extracellular miRNAs released from living PC cells accurately. The well-defined PNI nanostructure which provides a highly reproducible SERS hot spot line, straightforward probe LNA immobilization, and simple miRNA–LNA hybrid formation with equalized stabilities seems to collectively contribute to the observed equally enhanced and highly reproducible SERS signals for miR141 and miR375.
Conclusion
We have developed a PNI sensor that can detect extracellular miR141 and miR375 released from the cultured cells of PC cell lines. The proposed PNI sensor exhibited a low detection limit of 100 aM, a wide dynamic range from 100 aM to 100 pM, and a perfect discrimination of single-base mismatches in target miRNAs. By using the PNI sensor, we were able to estimate the absolute amount of the released miR141 and miR375 from each PC cell line. The highly sensitive and exactly quantifiable PNI sensor could be useful for the precise diagnosis of PC patients and will be further valuable for detecting other disease-related extracellular miRNAs.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016; 66: 7–30.
2. Moyer VA. Screening for prostate cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med 2012; 157: 120–134.
3. Cheng Z, Choi N, Wang R, Lee S, Moon KC, Yoon S-Y, Chen L, Choo J. Simultaneous detection of dual prostate specific antigens using surface-enhanced Raman scattering-based immunoassay for accurate diagnosis of prostate cancer. ACS Nano 2017; 11: 4926–4933.
4. Gupta SC, Tripatchi YN. Potential of long non-coding RNAs in cancer patients: from biomarkers to therapeutic targets. Int J Cancer 2017; 140: 1955–1967.
5. Kang T, Kim H, Lee JM, Lee H, Choi Y-S, Kang G, Seo M-K, Chung BH, Jung Y, Kim B. Ultra-specific zeptomole microRNA detection by plasmonic nanowire interstice sensor with bi-temperature hybridization. Small 2014; 10: 4200–4206.
6. Yang S, Kim H, Lee KJ, Hwang SG, Lim E-K, Jung J, Lee TJ, Park H-S, Kang T, Kim B. Attomolar detection of extracellular microRNAs released from living prostate cancer cells by a plasmonic nanowire interstice sensor. Nanoscale 2017; 9: 17387–17395.
7. Mohanty P, Yoon I, Kang T, Seo K, Varadwaj KSK, Choi W, Park Q-H, Ahn JP, Suh YD, Ihee H, Kim B. Simple vapor phase synthesis of single-crystalline ag nanowires and single nanowire surface-enhanced Raman scattering. J Am Chem Soc 2007; 129: 9576–9577.
8. Yoon I, Kang T, Choi W, Kim J, Yoo Y, Joo S-W, Park Q-H, Ihee H, Kim B. Single nanowire on a film as an efficient SERS-active platform. J Am Chem Soc 2009; 131: 758–762.
9. Kang T, Yoon I, Kim J, Ihee H, Kim B. Au nanowire-Au nanoparticles conjugated system which provides micrometer size molecular sensors. Chem Eur J 2010; 16: 1351–1355.
10. Kang T, Yoo SM, Kim B, Lee SY. Detection of single nucleotide polymorphism by a gold nanowire-on-film SERS sensor coupled with S1 nuclease treatment. Chem Eur J 2011; 17: 8657–8662.
11. Kang T, Yoo SM, Kang H, Lee H, Kang M, Lee SY, Kim B. Combining a nanowire SERRS sensor and a target recycling reaction for ultrasensitive and multiplex identification of pathogenic fungi. Small 2011; 7: 3371–3376.
12. Kang T, Yoo SM, Kang M, Lee H, Kim H, Lee SY, Kim B. Single-step multiplex detection of toxic metal ions by Au nanowires-on-chip sensor using reporter elimination. Lab Chip 2012; 12: 3077–3081.
13. Gwak R, Kim H, Yoo SM, Lee SY, Lee G-J, Lee M-K, Rhee C-K, Kang T, Kim B. Precisely determining ultralow level UO22+ in natural water with plasmonic nanowire interstice sensor. Sci Rep 2016; 6: 19646.
14. Lee JM, Hwang A, Choi HJ, Jo Y, Kim B, Kang T, Jung Y. A multivalent structure-specific RNA binder with extremely stable target binding but reduced interactions to nonspecific RNAs. Angew Chem Int Ed 2017; 56: 15998–16002.
15. Eom G, Kim H, Hwang A, Son H-Y, Choi Y, Moon J, Kim D, Lee M, Lim E-K, Jeong J, Huh Y-M, Seo M-K, Kang T, Kim B. Nanogap-rich Au nanowire SERS sensor for ultrasensitive telomerase activity detection: application to gastric and breast cancer tissue diagnosis. Adv Funct Mater 2017; 27: 1701832.
The authors
Taejoon Kang*1 PhD, Bongsoo Kim*2 PhD
1Hazards Monitoring Bionano Research Center, KRIBB, Daejeon 34141, Republic of Korea
2Department of Chemistry, KAIST, Daejeon 34141, Republic of Korea
*Corresponding author
E-mail: kangtaejoon@kribb.re.kr;
bongsoo@kaist.ac.kr