Blood-based tests for colorectal cancer screening
Worldwide, screening has been shown to reduce mortality and incidence of colorectal cancer. Despite its documented success, people still fail to participate and screening rates remain low in most countries. Given that patient-reported barriers include resistance to recommended fecal-based methods or endoscopy, blood-based tests have the potential to increase participation in colorectal cancer screening programmes.
by Dr Theo deVos
Background
Globally, colorectal cancer (CRC) is the third most common cancer in men and the second in women, with an estimated 1.36 million cases and causing an estimated 694,000 deaths in 2012 [1]. These rates are unnecessarily high since CRC is an excellent candidate for screening as evidenced by large randomized trials demonstrating reductions in mortality and incidence [reviewed in 2, 3]. Biologically, CRC usually develops slowly, going through a progression from non-cancerous polyp to cancer over a period of a decade or more. This biology readily lends itself to screening and early detection which has a significant positive impact on the effectiveness of intervention. For example, in the United States, 5-year survival is ~90 % if the tumour is confined locally when detected, ~70% if it has spread regionally, but only ~10% if distant metastases are present [4].
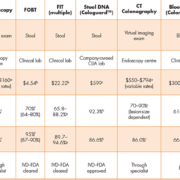
Colonoscopy is the predominantly recommended method for routine screening in some countries including the United States, as it enables detection and intervention in the same procedure. It is also the diagnostic follow-up for positive results of other screening tests. However, challenges with capacity and quality, financial concerns, and patient resistance have led to its lack of use as the primary screening modality in most settings. In some countries, flexible sigmoidoscopy is showing a resurgence, with reports demonstrating mortality and incidence benefits [2]. Table 1 displays a list of common CRC screening methods along with new methods coming on-line, today.
The first non-invasive tests for CRC were based on the detection of fecal occult blood (FOBT), and these have been further developed into immunological tests (FIT) using specific antibodies to detect hemoglobin. These tests are typically designed to allow patients to collect stool samples at home and ship the sample by mail to a central laboratory for testing. A newer alternative to fecal blood testing is the analysis of genetic/epigenetic markers in fecal material. This is the basis for the Cologuard test (Exact Sciences, WI, USA), a fecal DNA test recently approved by the US FDA [5]. Blood-based screening tests that measure tumour biomarkers in plasma or serum have been developed as a minimally-invasive alternative to fecal testing. DNA methylation tests based on SEPT9 have become available in Europe and are undergoing regulatory review in China. In addition, methylated SEPT9 testing is available as laboratory-developed tests (LDTs) in the USA, and a kitted version (Epi proColon®; Epigenomics AG, Germany) is currently undergoing US FDA premarket (PMA) review [6]. Another blood-based test, the ColonSentry risk test based on an expression panel is available as an LDT in the USA and in Japan.
Given the clear benefit of screening and the long standing availability of tests, the lack of participation is disappointing, and improving screening rates is a broadly accepted goal. As an example, the ‘80 by 2018’ campaign in the USA has set a goal of 80% adherence to screening guidelines by 2018 [7]. In order to meet this goal, barriers that prevent screening must be understood and overcome. There are numerous reports focused on understanding patient barriers to CRC screening. Although this is a complex issue involving costs, time, physician recommendation and several other factors, one consistent message from these studies is that the test methods themselves present barriers. Many patients are uncomfortable with all or part of the colonoscopy process and many are also uncomfortable with collecting and shipping fecal samples [8]. As a consequence, CRCs are diagnosed symptomatically in more instances than necessary, when the disease has spread beyond the primary site, resulting in greatly reduced survival rates. The availability of a screening test using a simple and common blood draw, which can be included as part of a regular check-up, has the potential to overcome some barriers and improve screening rates.
Blood-based screening
There are a number of approaches to the measurement of cancer biomarkers in the blood. The detection and quantification of circulating tumour cells represents an early approach, which was developed into a commercial system (e.g. CellSearch; Janssen Diagnostics, NJ, USA) though this analysis has not generally been used for cancer screening. Another alternative derives from the isolation and fractionation of circulating immune cells and the quantification of gene expression panels correlated with the disease by reverse-transcriptase PCR. This ‘sentinel concept’ is the basis for the ColonSentry test (GeneNews, Canada) in Table 1. A third alternative is the measurement of metabolic products by mass-spectrometry that are correlated with the presence of cancer. As an example, a commercial test (Cologic; Phenomenome, Canada) was developed based on the measurement of serum levels of GTA-446, an anti-inflammatory fatty acid. The most developed and perhaps simplest approach in this field is the measurement of cell-free genetic or epigenetic markers in plasma or serum that are highly correlated with the presence of cancer. As shown in Table 1, the methylated Septin9 biomarker and the Epi proColon® test were developed based on this approach.
Screening biomarkers in plasma and serum
The recognition that tumour DNA contains genetic and epigenetic changes that can serve as biomarkers dates back a number of decades. As reviewed recently, the list of biomarker reports for colorectal cancer grows ever longer [9]. Although numerous studies report on marker performance, the majority of studies include only a limited number of cases and controls, and only a small subset of markers have been rigorously tested in the clinical setting. Furthermore, a review of marker studies in ClinicalTrials.gov indicated very few ongoing CRC marker screening trials. Well validated markers include methylated SEPT9 described above, and the methylation of BCAT1 and IKZF1 sequences in plasma which have shown to be correlated with CRC [10] and are currently being tested in a clinical trial in Australia. There are many interesting genetic and epigenetic markers, but most await additional validation data that will support clinical utility.
Laboratory considerations for a plasma-based screening test
The basic concept outlined in Figure 1 illustrates key points associated with development of a genetic/epigenetic screening test. CRC screening from blood samples imposes rigorous demands that impact the reduction to practice for a test including: (a) high volume (millions of tests); (b) low target copy number (~1 copy per mL); (c) fragmented DNA; (d) large sample size (e.g. 3.5 mL); and (e) kitted reagents. These are discussed using the methylated Septin9 test as a case study.
Blood draw and processing
Given that screening is a high volume activity, an inexpensive and standard sample collection method is beneficial. In this case, a simple blood draw using a standard collection tube (e.g. K2EDTA plasma collection tube) is performed at the clinic or draw station. Plasma or serum is separated and if necessary they can be re-centrifuged to ensure cell-free status. The emphasis is on preparing cell-free material to limit background contamination due to lysis of nucleated cells in the blood. While this has led to the use of specialized collections tubes (Streck, NE, USA) in the field of prenatal diagnostics, these have not been widely tested for colorectal cancer screening. Cleared plasma can be tested immediately, or stored frozen for a period of time.
Nucleic acid extraction
In this step, cell-free nucleic acids are extracted from the plasma sample. While a number of commercial methods have been developed for this purpose, it remains the Achilles heel of the process. Given the wide range in target concentration, and particularly the exceptionally low copy number expected for early cancers (in the single copy per mL range) [6], as well as the fragmented nature of cell-free DNA, the extraction methods must be designed to handle large samples (e.g. 3–4 mL of plasma), and be able to isolate fragmented DNA. The use of magnetic particles for purification coupled with modified binding and wash buffers designed to capture the full range of DNA fragments has simplified the extraction, and with the development of liquid handling platforms that can process larger volumes, this step is becoming automatable. While the reduction from 3.5 mL plasma to 100 µL of DNA eluate would raise concerns for PCR inhibition, for DNA methylation tests, it is possible to reduce the wash steps because the DNA is extensively purified in the bisulfite treatment process.
Bisulfite treatment
The bisulfite treatment process is required if the target is DNA methylation-based. Recent improvements in bisulfite conversion technology have simplified the treatment. The change to ammonium bisulfite allows for liquid reagents – a key attribute for kit development. In combination with elevated temperatures, bisulfite incubation time is reduced to less than 1 hour, enabling single shift turn-around times for tests. Furthermore, the reaction can be purified using a magnetic particle extraction that takes advantage of the same particles used for the initial DNA extraction. This process can also be automated on a standard liquid handling platform to improve throughput and quality.
Real-time PCR
For genetic (mutation)-based tests, the test can be performed immediately following initial DNA extraction, though it is important to increase the stringency of DNA washes to limit the potential for PCR inhibition. In the final steps, either genetic or epigenetic markers are measured by real-time PCR. For screening applications, the target concentration dictates the conditions and interpretation of the PCR reaction. For example, in the methylated Septin9 test, the final recovered bisulfite converted template DNA is split into three wells and run in three PCR reactions. Although the PCR reaction is run as a real-time assay, the test is essentially a qualitative end point test, since a well is called positive if a PCR curve occurs at any cycle during the course of the reaction. In addition, the results of the three reactions are combined to produce a final interpretation for a patient sample. For the CE-marked Epi proColon 2.0 product, the sample is called positive if two of three wells are positive. For the Ep proColon product undergoing US FDA PMA review, the sample is called positive if any of three wells are positive. This allows for a greater emphasis on a specific test parameter – for sensitivity (any well-positive) or test specificity (two out of three wells positive).
Summary
The use of genetic and epigenetic biomarkers for cancer screening is a field still in its infancy that has great opportunities for growth. Because these biomarkers can be used as indicators of disease, they also have diagnostic and prognostic potential that will be incorporated into the clinical-decision making process. For CRC screening, test kits are already available in Europe and other countries, and are currently under review by both the US and Chinese FDA organizations. In the US, LDTs are currently marketed, and together, all progress represents significant opportunities to generate positive momentum. The introduction of simple, blood-based screening would provide a viable alternative to patients refusing or avoiding current well established methods. The convenience factors of sample collection and processing by health professionals also avoids the challenges of faulty sampling, handling, and mailing associated with at-home self-collected tests. Finally, given the extensive collection of promising biomarkers on the horizon, mechanisms are needed now to expedite clinical utilization and validation to drive further improvements in test performance.
References
1. Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray, F. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet]. Lyon, France: International Agency for Research on Cancer; 2013. Available from: http://globocan.iarc.fr, accessed on 12/09/2014.
2. Kuipers EJ, Rösch T, Bretthauer M. Colorectal cancer screening – optimizing current strategies and new directions. Nat Rev Clin Oncol. 2013; 10: 130–142.
3. Brenner H, Stock C, Hoffmeister M. Effect of screening sigmoidoscopy and screening colonoscopy on colorectal cancer incidence and mortality: systematic review and meta-analysis of randomised controlled trials and observational studies. BMJ 2014; 348: g2467.
4. American Cancer Society. Colorectal Cancer Facts & Figures 2014-2016. Atlanta: American Cancer Society, 2014.
5. Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, Ahlquist DA, Berger BM. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014; 370(14): 1287–1297.
6. Potter NT, Hurban P, White MN, Whitlock KD, Lofton-Day CE, Tetzner R, Koenig T, Quigley NB, Weiss G. Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin Chem. 2014; 60(9): 1183–1191.
7. National Colorectal Cancer Round Table. Tools & Resources – 80% by 2018. http://nccrt.org/about/80-percent-by-2018/
8. Gimeno García AZ. Factors influencing colorectal cancer screening participation. Gastroenterol Res Pract. 2012; 2012: 483417.
9. Toiyama Y, Okugawa Y, Goel A. DNA methylation and microRNA biomarkers for noninvasive detection of gastric and colorectal cancer. Biochem Biophys Res Commun. 2014; doi: 10.1016/j.bbrc.2014.08.001.
10. Mitchell SM, Ross JP, Drew HR, Ho T, Brown GS, Saunders NF, Duesing KR, Buckley MJ, Dunne R, Beetson I, Rand KN, McEvoy A, Thomas ML, Baker RT, Wattchow DA, Young GP, Lockett TJ, Pedersen SK, Lapointe LC, Molloy PL. A panel of genes methylated with high frequency in colorectal cancer. BMC Cancer 2014; 14: 54.
The author
Theo deVos PhD
Epigenomics Inc.,
Seattle, WA 98107, USA
E-mail: theo.devos@epigenomics.com