Chinese academy develops AI-powered protein engineering platform
Chinese researchers have developed AiCE, an artificial intelligence-informed approach that dramatically enhances protein engineering efficiency. The method integrates structural and evolutionary constraints into existing AI models, achieving 36-90% improvements in predictive accuracy whilst eliminating the need for specialised model training.
The Chinese Academy of Sciences has unveiled a transformative approach to protein engineering that promises to accelerate biotechnology applications across precision medicine and molecular breeding. The AI-informed Constraints for protein Engineering (AiCE) platform, developed by Professor Gao Caixia’s team at the Institute of Genetics and Developmental Biology, represents a significant advancement in computational protein design methodology.
Published in Cell, the research addresses fundamental limitations in current protein engineering approaches, which often suffer from high computational costs, limited scalability, and accessibility barriers that restrict widespread adoption across research communities.
Enhanced prediction accuracy through structural integration
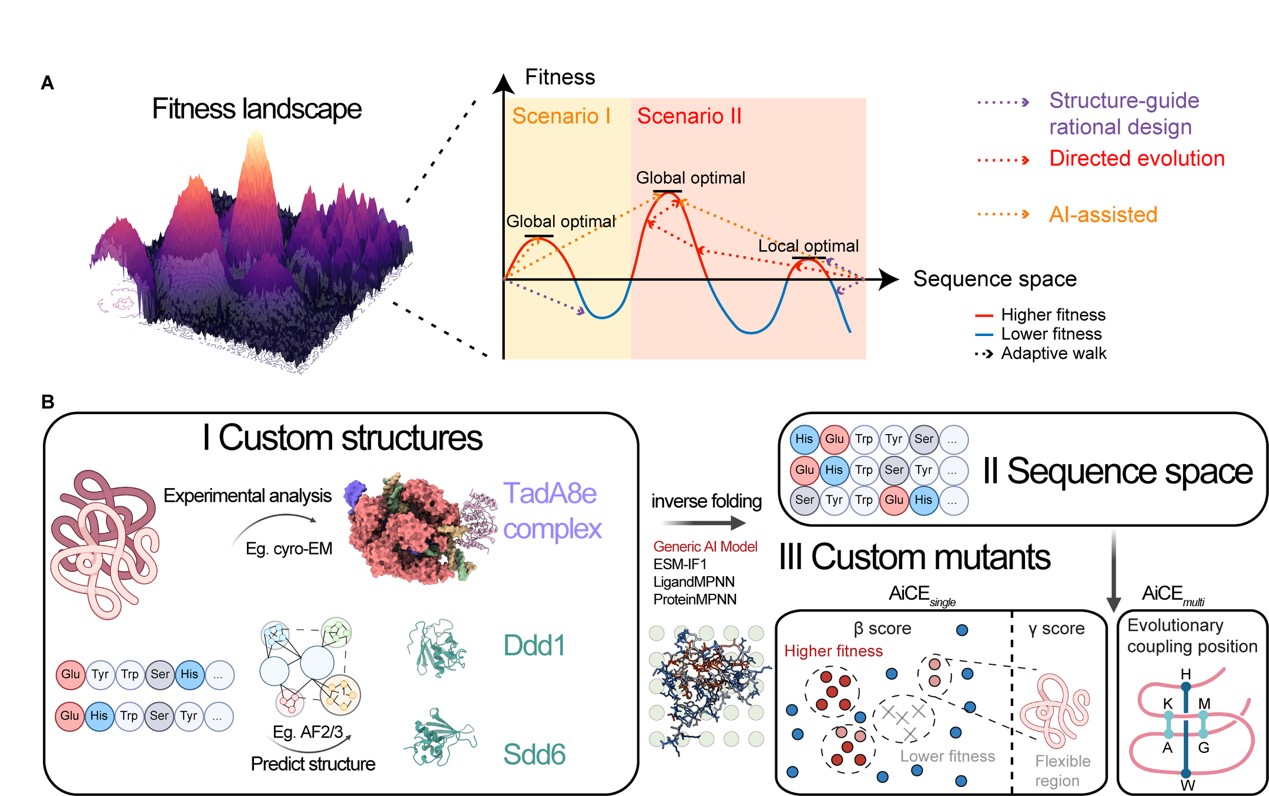
The AiCE framework comprises two complementary modules designed to optimise protein function prediction. The AiCEsingle component focuses on identifying high-fitness single amino acid substitutions by extensively sampling inverse folding models – sophisticated AI systems that generate compatible amino acid sequences based on three-dimensional protein structures.
Benchmarking studies conducted across 60 deep mutational scanning datasets demonstrated AiCEsingle’s superior performance, outperforming existing AI-based methodologies by 36-90%. The incorporation of structural constraints alone yielded a 37% improvement in predictive accuracy, validating the approach’s effectiveness for complex proteins and protein-nucleic acid complexes.
Addressing combinatorial mutation challenges
To tackle negative epistatic interactions in combinatorial mutations, the research team developed AiCEmulti, which integrates evolutionary coupling constraints. This module enables accurate prediction of multiple high-fitness mutations whilst maintaining minimal computational overhead, significantly expanding the platform’s practical utility.
The researchers successfully demonstrated AiCE’s capabilities by engineering eight proteins with diverse structural and functional characteristics, including deaminases, nuclear localisation sequences, nucleases, and reverse transcriptases.
Clinical applications and next-generation editors
The AiCE platform has facilitated the creation of several advanced base editors with enhanced therapeutic potential. Notable developments include enABE8e, a cytosine base editor featuring a 50% narrower editing window for improved precision, enSdd6-CBE, an adenine base editor with 1.3-fold higher fidelity, and enDdd1-DdCBE, a mitochondrial base editor demonstrating a 13-fold increase in activity.
These engineered proteins address critical requirements in precision medicine applications, where enhanced specificity and reduced off-target effects are paramount for clinical translation.
Democratising protein engineering capabilities
The AiCE methodology represents a paradigm shift towards accessible protein engineering solutions. By leveraging existing AI models rather than requiring specialised training protocols, the approach reduces computational barriers whilst maintaining predictive accuracy.
The framework’s interpretability features provide researchers with clearer insights into AI-driven protein redesign processes, facilitating more informed experimental design and validation strategies. This transparency addresses longstanding concerns regarding the “black box” nature of AI-based protein engineering approaches.
The research establishes a foundation for broader applications in biotechnology, pharmaceutical development, and agricultural innovation, where enhanced protein function prediction capabilities could accelerate product development timelines and improve therapeutic outcomes.
Reference
Gao, C., et al. (2025). Enhanced protein evolution with inverse folding models using structural and evolutionary constraints. Cell, 188(14). https://doi.org/10.1016/j.cell.2025.06.014