Colistin resistance detection in Acinetobacter baumannii by mass spectrometry of microbial lipids
Acinetobacter baumannii is a prevalent nosocomial pathogen with a high incidence of multidrug resistance. Treatment of infections with colistin can result in emergence of colistin-resistant strains. This occurs via modifications of the phosphate moieties of lipopolysaccharide-derived lipid A, which are readily identified by mass spectrometry (MS). In this article, we describe colistin susceptibility determinations by lipid MS of A. baumannii and our recent study in which we correlate MS results with traditional antimicrobial susceptibility testing of clinical isolates.
by Dr Lisa M. Leung, Dr Robert A. Myers, Dr Yohei Doi and Prof. Robert K. Ernst
Background
Colistin resistance in Gram-negative pathogens
Multidrug-resistant, Gram-negative bacterial pathogens continue to pose serious threats to public health. Carbapenem-resistant Enterobacteriaceae (CRE), Pseudomonas aeruginosa (CRPA), and Acinetobacter baumannii (CRAB) are given the highest global priority among drug-resistant organisms by organizations, such as the World Health Organization (WHO) and the Centers for Disease Control and Prevention (CDC) [1]. Carbapenem-resistant infections can be treated with colistin, a last resort antibiotic of the polymyxin class, leading to an increase in colistin resistance and resulting in devastating consequences as it is one of the last remaining effective antimicrobials [2]. Furthermore, discovery of a plasmid-mediated colistin resistance gene, mcr, has intensified this urgency given the potential for rapid and widespread dissemination of colistin-resistant bacteria across the globe [3, 4]. Therefore, the WHO and CDC have prioritized development of novel diagnostics and therapeutics to address the global threat of pathogens, such as multidrug-resistant A. baumannii [5].
A novel diagnostic approach is proposed
In elucidating the mechanism of colistin resistance, researchers analysed microbial glycolipids by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). These findings contributed to determination of the resistance mechanism in A. baumannii, via addition of phosphoethanolamine onto the terminal phosphate moieties of the lipopolysaccharide (LPS)-derived lipid A (LA), decreasing the electronegativity of the membrane and, subsequently, the binding affinity of colistin [6]. These modifications create unique features on the resultant mass spectra of colistin-resistant strains that can be used as a diagnostic marker. Our group has published proof-of-concept studies utilizing this platform in the identification of the ESKAPE pathogens [7], as well as elucidation of colistin susceptibility in organisms such as Klebsiella pneumoniae [8], E. coli, and P. aeruginosa [9]. Protein-based microbial identification using MALDI-TOF MS is a simple and effective means of identifying causative agents although it still faces challenges, such as identification of closely related organisms (Candida or Shigella subspecies), antimicrobial susceptibility determination, or identification of organisms in polymicrobial or biologically relevant samples (urine, blood or wound effluent) [10]. Therefore, we offered this novel platform as an alternative and complementary approach to strengthen the overall diagnostic power of MALDI-TOF MS and continue to demonstrate its capability in our latest study detecting colistin resistance in A. baumannii [11].
Methods and results
Overview of clinical data
In this study, we prospectively collected A. baumannii complex clinical isolates from a hospital system in Pennsylvania between 2014 and 2016, a total of 451 isolates from 284 patients. Among the 284 unique isolates from each patient, 73.6% (209 isolates) were determined to be A. baumannii, 18.7% (53 isolates) Acinetobacter pittii, 3.5% (10 isolates) Acinetobacter nosocomialis, and 1.8% (5 isolates) Acinetobacter calcoaceticus. The remaining <1% were identified as the following Acinetobacter genospecies that do not belong to the A. baumannii complex: Acinetobacter radioresistens (2 isolates), Acinetobacter guillouliae (1 isolate), and Acinetobacter junii (1 isolate). Three isolates (0.7%) could not be reliably identified. All isolates were evaluated for colistin resistance using standard minimum inhibitory concentration (MIC) testing by both agar dilution and broth microdilution in accordance with the clinical breakpoint provided by the EUCAST [12]. Of the 451 clinical isolates, 394 isolates from 249 patients were found to be susceptible to colistin (≤2 µg/mL), and a total of 39 isolates (8.6%) from 20 patients were identified as resistant (>2 µg/mL).
The colistin-resistant A. baumannii mass spectrum
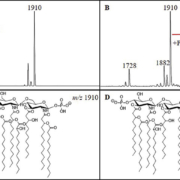
All strains were cultured overnight and subjected to a hot ammonium isobutyrate reaction to extract cellular lipids. Extracts were analysed by MALDI-TOF in negative ion mode using a Bruker microflex LRF MALDI-TOF mass spectrometer operated in reflectron mode and using norharmane as a matrix. Ions most often observed in the mass spectra were m/z 1404, 1728, and 1910; these have been previously characterized [6], with m/z 1910 representing the full bis-phosphorylated, hepta-acylated lipid A structure (Fig. 1). Resistant isolates were defined by the presence of an ion at m/z 2033, representing the addition of a phosphoethanolamine moiety to one of the phosphate moieties of the m/z 1910 structure (∆m/z=123) (Fig. 1). Determination of resistance was made by observing this ion in acquired mass spectra for each sample above a signal-to-noise ratio of 3. Of the 451 clinical isolates, 397 were determined to be susceptible to colistin (i.e. lacking an ion at m/z 2033), whereas 54 (12.0%) showed the presence of the m/z 2033 and were classified as resistant.
Differentiation of the A. baumannii complex
Differences were observed between spectra collected from the A. baumannii complex isolates, A. baumannii, A. pittii, and A. nosocomialis (Fig. 2). In general, an ion at m/z 1882 displayed higher signal intensity in A. pittii and A. nosocomialis isolates, about 80% relative intensity to the base peak at m/z 1910 compared to about 10% for A. baumannii, which may indicate differences in relative abundances of specific LPS structures. This ion most likely results from an exchange of a shorter chain fatty acyl group (C2H4, ∆m/z=28) from one of the acyl chains of the base structure at m/z 1910, although this structure is inferred and further analyses would need to be conducted for positive structural determinations. In addition, A. pittii and A. nosocomialis isolates showed prominent novel ions at m/z 1866 and 1894, indicating differences in hydroxylation events (∆m/z=16) from ions at m/z 1882 and 1910, respectively, potentially representing the addition of a hydroxyl moiety to one of the attached fatty acyls of lipid A. Among the 39 colistin-resistant isolates, only one was identified as non-baumannii (A. nosocomialis). This means that non-baumannii isolates occur at a lower incidence among resistant isolates (3.1%), as compared to their incidence among Acinetobacter isolates in general (19.7%) indicating a higher resistance rate of A. baumannii versus non-baumannii complex isolates in this study.
MIC versus MS
Discordant results between MIC and MS findings were resolved by multiple-replicate retesting to confirm susceptibility profiles, and final determinations were compared. Of the 451 total isolates used in our study, 394 isolates from 249 patients were determined to be susceptible by both MIC and MS and 39 isolates from 20 patients were determined to be resistant, giving a specificity of 94.0% and a sensitivity of 92.9%. Three isolates were determined to be resistant by MIC yet susceptible by MS and 15 isolates were found to be resistant by MS but susceptible by MIC. When considering only the first isolates isolated from the 284 patients in our study, sensitivity and specificity values change slightly – to 83.3% and 97.4%, respectively. Thirty-nine isolates were subjected to multiple-replicate retesting based on discordant results between agar dilution and broth microdilution methods, MIC and MS results, or both. Of the 33 isolates that underwent MIC retesting, 26 (or 89.7%) gave different susceptibility profiles, 25 went from resistant to susceptible and one was classified as indeterminate. Of the 26 isolates that underwent MS retesting, only three (11.5%) saw a change in their susceptibility profiles; two went from resistant to susceptible and one from susceptible to resistant. Although there was a high association between susceptibility determinations by MIC and MS overall, the positive predictive value was calculated as 72.2% (negative predictive value=99.2%). This is largely owing to the 15 isolates where resistance-associated ions were observed in the mass spectra, but which were determined susceptible by MIC. Chromosomally-mediated colistin resistance in Acinetobacter species is due to overexpression of LPS-modifying genes; therefore, modification of LPS will vary over time. It is presently unclear whether this ‘resistant’ profile is a valid determination of resistance or whether this isolate would present as a resistant infection in a clinical scenario.
Conclusion
A. baumannii, a prevalent, Gram-negative coccobacillus pathogen, poses a significant challenge to clinicians due to the incidence of hospital-acquired and drug-resistant infections. Close monitoring of this pathogen and other A. baumannii complex organisms is considered of critical importance to public health organizations. Here, we surveyed 451 Acinetobacter isolates prospectively collected from patients at a major Pennsylvania health system over a 3-year period. We determined colistin resistance by MIC testing, as well as by MALDI-TOF MS. As in previous studies of colistin-resistant K. pneumoniae, P. aeruginosa, and A. baumannii [6, 8, 13], the data showed a strong association between resistant MIC determinations and the observation of higher m/z ions by MS consistent with modification to LA and previously demonstrated to confer resistance. A. nosocomialis, A. pittii, and A. calcoaceticus, along with A. baumannii are collectively identified as the A. baumannii complex organisms. In our prospective study, we found that A. baumannii isolates were the predominant species within the A. baumannii complex, yet represented a smaller proportion (73.6%) than what has previously been observed [14]. We also demonstrated that a lipid MS profile offers another diagnostic tool for differentiation and accurate surveillance of these pathogens. Furthermore, the finding of resistance ions among a resistant A. nosocomialis isolate demonstrates that A. baumannii complex organisms likely achieve colistin resistance via the same LPS-modifying mechanism (Fig. 2). Overall, we conclude that glycolipid MS profiling can effectively detect colistin resistance in A. baumannii and has the potential to direct antimicrobial stewardship in the clinic, further validating our recently introduced diagnostic platform [7].
References
1. Antibiotic resistance threats in the United States, 2013; p114. Centers for Disease Control and Prevention 2013 (https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf)
2. Osei Sekyere J, Govinden U, Bester LA, Essack SY. Colistin and tigecycline resistance in carbapenemase-producing Gram-negative bacteria: emerging resistance mechanisms and detection methods. J Appl Microbiol 2016; 121(3): 601–617.
3. Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 2016; 16(2): 161–168.
4. Bardet L, Rolain JM. Development of new tools to detect colistin-resistance among Enterobacteriaceae strains. Can J Infect Dis Med Microbiol 2018; 2018: 3095249.
5. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organization 2017 (https://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/).
6. Pelletier MR1, Casella LG, Jones JW, Adams MD, Zurawski DV, Hazlett KR, et al. Unique structural modifications are present in the lipopolysaccharide from colistin-resistant strains of Acinetobacter baumannii. Antimicrob Agents Chemother 2013; 57(10): 4831–4840.
7. Leung LM, Fondrie WE, Doi Y, Johnson JK, Strickland DK, Ernst RK, et al. Identification of the ESKAPE pathogens by mass spectrometric analysis of microbial membrane glycolipids. Sci Rep 2017; 7(1): 6403.
8. Leung LM, Cooper VS, Rasko DA, Guo Q, Pacey MP, McElheny CL, Mettus RT, et al. Structural modification of LPS in colistin-resistant, KPC-producing Klebsiella pneumoniae. J Antimicrob Chemother 2017; 72(11): 3035–3042.
9. Liu YY, Chandler CE, Leung LM, McElheny CL, Mettus RT, Shanks RMQ, et al. Structural modification of lipopolysaccharide conferred by mcr-1 in Gram-negative ESKAPE pathogens. Antimicrob Agents Chemother 2017; 61(6): pii: e00580-17.
10. Elssner T, Kostrzewa M, Maier T, Kruppa G. Microorganism identification based on MALDI-TOF-MS fingerprints. In NATO Science for Peace and Security Series A: Chemistry and Biology, pp. 99–113. Springer 2011.
11. Leung LM, McElheny CL, Gardner FM, Chandler CE, Bowler SL, Mettus RT, et al. A prospective study of Acinetobacter baumannii complex isolates and colistin susceptibility monitoring by mass spectrometry of microbial membrane glycolipids. J Clin Microbiol 2019; 57(3): pii: e01100-18.
12. Recommendations for MIC determination of colistin (polymyxin E ); as recommended by the joint CLSI-EUCAST Polymyxin Breakpoints Working Group. EUCAST 2016 (http://www.bioconnections.co.uk/files/merlin/Recommendations_for_MIC_determination_of_colistin_March_2016.pdf).
13. Miller AK, Brannon MK, Stevens L, Johansen HK, Selgrade SE, Miller SI, et al. PhoQ mutations promote lipid a modification and polymyxin resistance of Pseudomonas aeruginosa found in colistin-treated cystic fibrosis patients. Antimicrob Agents Chemother 2011; 55(12): 5761–579.
14. Queenan AM, Pillar CM, Deane J, Sahm DF, Lynch AS, Flamm RK, et al. Multidrug resistance among Acinetobacter spp. in the USA and activity profile of key agents: results from CAPITAL Surveillance 2010. Diagn Microbiol Infect Dis 2012; 73(3): 267–270
The authors
Lisa M. Leung1,2 PhD, Robert A. Myers3 PhD, Yohei Doi4 MD, and Robert K. Ernst*2 PhD
1Divisions of Molecular Biology and Microbiology, Maryland Department of Health Laboratories Administration, Baltimore, MD, USA
2Department of Microbial Pathogenesis, University of Maryland School of Dentistry, Baltimore, MD, USA
3Maryland Department of Health Laboratories Administration, Baltimore, MD, USA
4Division of Infectious Diseases, University of Pittsburgh School of Medicine, Pittsburgh, PA, USA
*Corresponding author
E-mail: rkernst@umaryland.edu