Exploring blood biomarkers for monitoring COVID-19 disease progression and outcomes and impact of SARS-CoV-2 variants of concern
In April 2020, the Royal College of Pathologists published recommendations for
a set of blood COVID-19 biomarkers to monitor disease severity and progression, which have since been employed by many UK hospital diagnostics services. Subsequent findings regarding biomarkers and their ability to distinguish or predict severe disease remain under investigation and emerging evidence highlights that interpretation may be influenced by different variants of concern.
Introduction
Since the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), there has been an unprecedented global effort to understand the pathogenesis and clinical progression of the disease it causes, coronavirus disease 2019 (COVID-19). Early evidence from China provided insight into several laboratory findings common in COVID-19 [1], and much of this formed the basis of the Royal College of Pathologists April 2020 guidance on the use and interpretation of clinical biochemistry tests in patients with SARS-CoV-2 infection [2]. In partnership with local clinical teams, Coventry and Warwickshire Pathology service (CWPS) were early adopters of this biomarker panel and made it available to all COVID-19 admissions; this required rapid implementation of assays for interleukin-6 (IL-6) and procalcitonin. Having this panel in routine clinical use for the care of COVID-19 inpatients from April 2020, has enabled the pathology service to obtain and interrogate a vast amount of biomarker data covering the various waves of the pandemic, each attributed to different variants of concern (VOCs).
COVID-19 pandemic timeline and emergence of variants
The occurrence of mutations in both RNA and DNA viruses during replication is a well-documented biological property crucial for fitness to survival. Genetic changes in key proteins that confer a competitive advantage (e.g. improved cell entry, ability to evade the host immune response or increased transmissibility) can be favoured via natural selection; allowing for an increased number of those virions that carry the mutation within the circulating population [3]. According to the World Health Organization (WHO), a variant is deemed of interest if one or more of the following are seen:
• an increase in transmissibility or detrimental change in epidemiology;
• an increase in virulence or change in clinical presentation; or
• a decrease in effectiveness of public health and social measures [4].
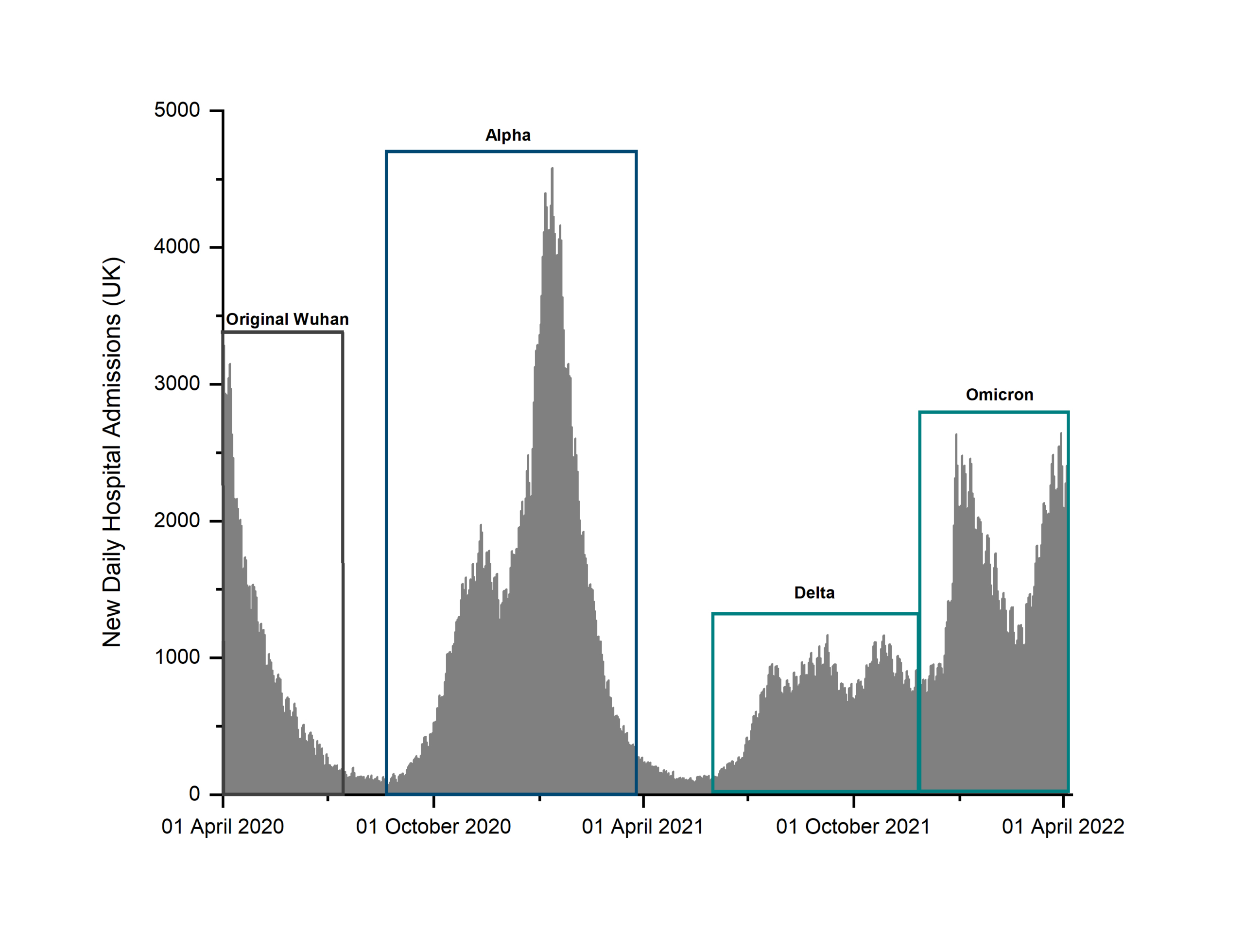
In the UK, the timeline of COVID-19 pandemic based on healthcare data such as hospital admissions can be broadly divided into three stages (Fig. 1): the early initial wave that peaked in mid-April 2020, a second bi-phasic phase with a prodromic peak in October–November 2020 followed by a much larger wave that peaked in mid-January 2021 associated with the Alpha variant, and a sub-
sequent third ‘tidal-like’ wave that started in the second half of 2021 [5]. This third stage has been sustained by the emergence of virus variants with increased transmissibility and infectivity, such as the delta and omicron.
Biomarkers and severity
SARS-CoV-2 enters the lungs through binding of the spike protein to the specific receptor angiotensin-converting enzyme 2 (ACE2), expressed by alveolar epithelial cells. Receptor expression has also been identified in cells found in the intestine, kidney, heart, adipose and reproductive tissues [6]. For those infected, up to 39% may be asymptomatic [7]. In patients with COVID-19 disease, symptoms are heterogeneous and range from a mild cough or fever to the more severe presentation seen in up to 10% of cases, with symptoms such as pneumonia, respiratory failure, sepsis, multiple organ dysfunction or death [7].
The underlying pathological processes behind these outcomes and why some individuals become more acutely unwell are not yet fully understood, although genetic and previous health history appear to be important. The more severe end of the spectrum has been associated with uncontrolled secretion of pro-inflammatory cytokines, resulting in a ‘cytokine storm’, leading to an increased risk of vascular hyperpermeability. This phenotype is often associated with thrombotic events, itself associated with hemostatic abnormalities, tissue damage in the heart, liver and kidney, as well as respiratory failure or multiple organ failure [8]. It has been proposed that the widespread distribution of the ACE2 receptor, endothelial damage and dysregulated immune response, could all be possible mechanisms for systemic organ involvement.
From the various published studies investigating biomarkers and outcomes of COVID-19 disease, current clinical data indicates that poor outcomes (evidenced by mechanical ventilation, critical care stay or death), are associated with uncontrolled inflammatory responses as seen through raised C-reactive protein (CRP), IL-6 and lactate dehydrogenase (LDH). Other common findings include lymphopenia with an increase in both neutrophil count and neutrophil-to-lymphocyte ratio, elevated levels of urea, creatinine and liver enzymes and decreased platelet counts [9]. Insight into specific biomarker profiles and how they differ according to patient outcome or time course of the disease could aid in the development or refinement of predictive markers, ultimately allowing for the stratification of patients and guided medical management or targeted therapy.
The B.1.1.7 (Alpha) variant
The B.1.1.7 variant, more commonly known as Alpha, changed the shape of the pandemic by exhibiting accelerated rates of transmissibility. Specifically, this variant accrued 23 mutations across the genome, with the two most important mutations identified as a non-synonymous mutation affecting the receptor binding domain of the spike protein at position 501 and a H69/V70 deletion, likely leading to a conformational change in the spike protein [10]. It was this variant that drove the second, most lethal wave of infections, which was characterized by increased hospitalization, disease severity and death rates, especially as the population vaccination rates were low at that time.
During the early stages of the Alpha-led wave, there was disagreement as to whether Alpha exhibited increased pathogenicity in comparison to earlier variants or whether enhanced transmission itself was responsible for increased incidence, hospital admissions and therefore deaths. Although population studies in early 2021 did not reveal any change in symptoms, disease duration or mortality [10], later cohort studies have since reported an increased hazard ratio for risk of death and observational studies have identified an increased risk of critical care admission and 28-day mortality [11].
CWPS COVID-19 surveillance protocol
Following conflicting reports and a paucity of studies analysing real-word routine data of hospitalized patients during the different waves of the COVID-19 pandemic, CWPS initiated a VOC surveillance programme that involved monitoring patterns of VOC spread and analysis of both extracted biomarker data and related information regarding clinical outcomes [12]. In the case of the Alpha-VOC-led wave, this involved analysis of samples and data between October 2020 and April 2021. This can be used to study changes and temporal dynamics of biomarkers during periods of interest and also identify differences in biomarker averages over time. We used this approach as a proxy for altered disease characteristics in hospitalized patients relevant to emergence of VOCs, such as the Alpha/B.1.1.7. Further refinement of analysis outcomes can be achieved by including patient characteristics and variables such as age, sex, ethnic group, bodymass index, comorbidities, smoking and social status. In a distinct approach, COVID-19 biomarker profiles from cohorts of patients with confirmed VOC status can be correlated with outcomes of hospital admission (death versus survival). This can be particularly useful when longitudinal data are available for individuals with multiple biomarker measures throughout the COVID-19 admission and hospitalization continuum.
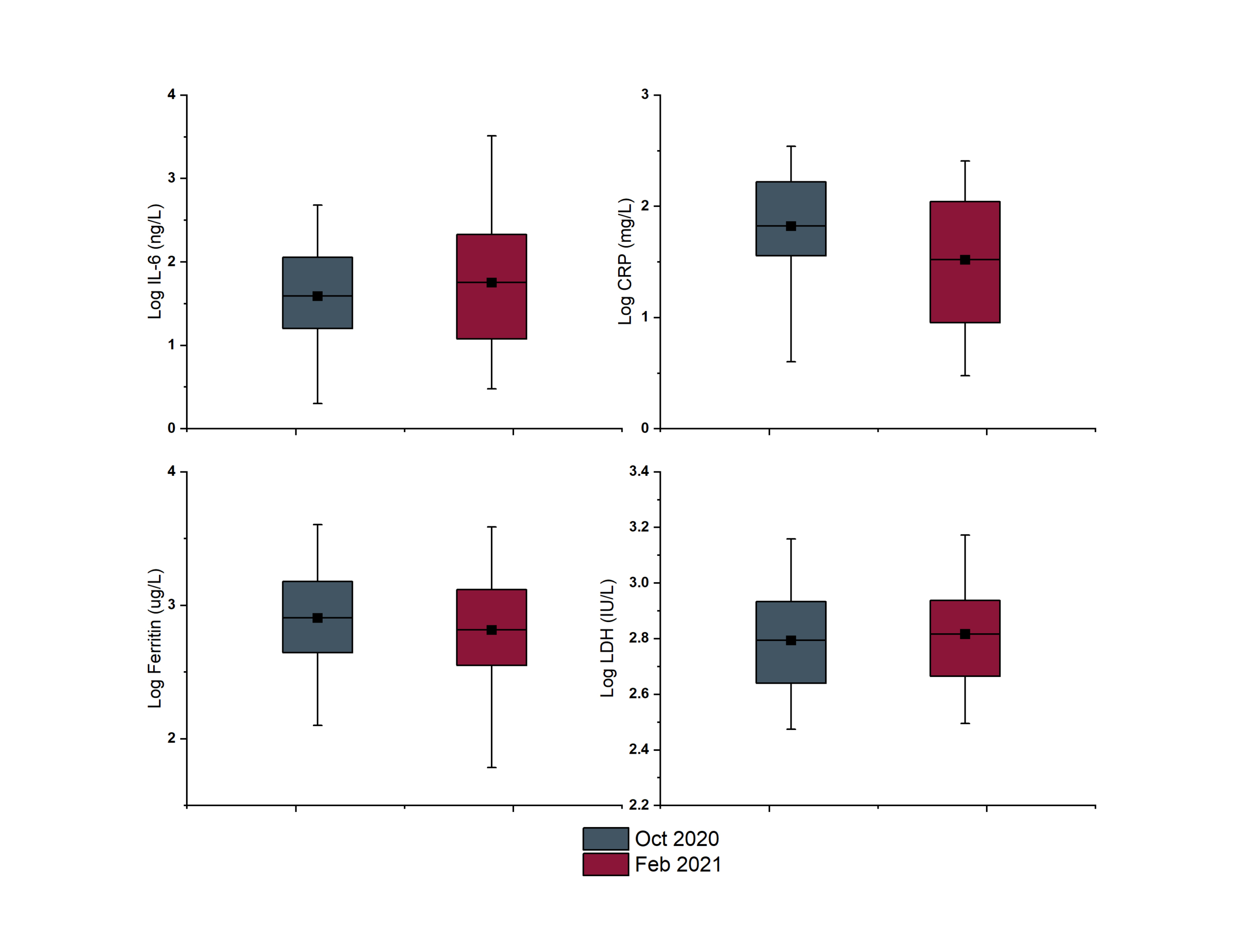
Across the study period, biomarkers exhibited distinct temporal dynamics, with most analytes within the biomarker panel being relatively stable. Small fluctuations were visible as peaks and troughs across the months for COVID-19 patients, whereas tighter distribution was seen for non-COVID patients. Interestingly, some biomarkers did exhibit quite significant changes over time. Figure 2 gives examples of such changes highlighted between two different months: October 2020 (when infections were generally attributed to wild-type SARS-CoV-2) and February 2021 (where the Alpha VOC was dominant within the local population). For the biochemistry markers ferritin and LDH, very little difference can be seen across the two time periods. However, for both IL-6 and CRP a change can be seen in the distribution of values and the mean. A wider spread of results was reported in the February period for both analytes, with the mean of IL-6 measured in COVID patients increasing and the mean CRP decreasing.
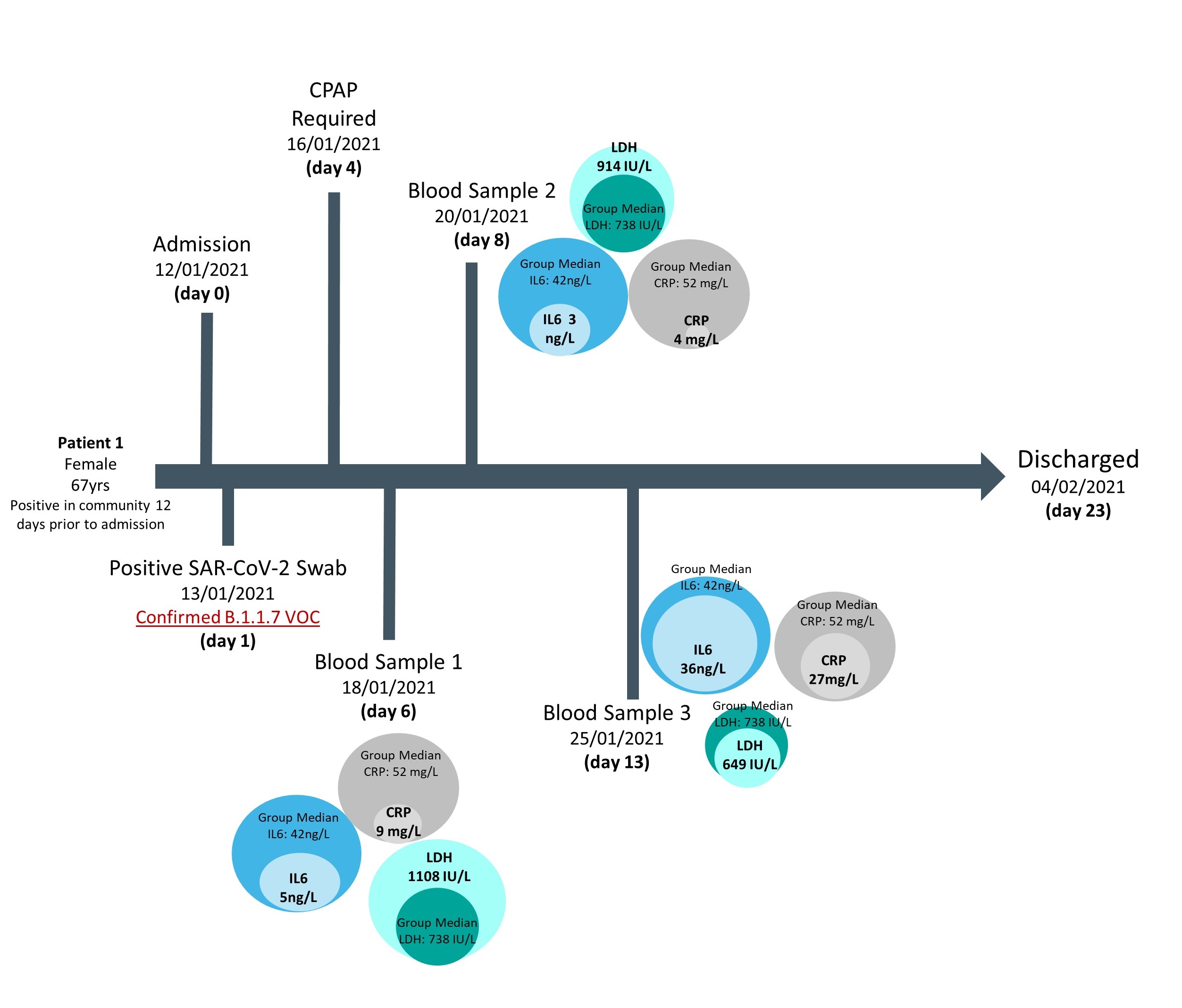
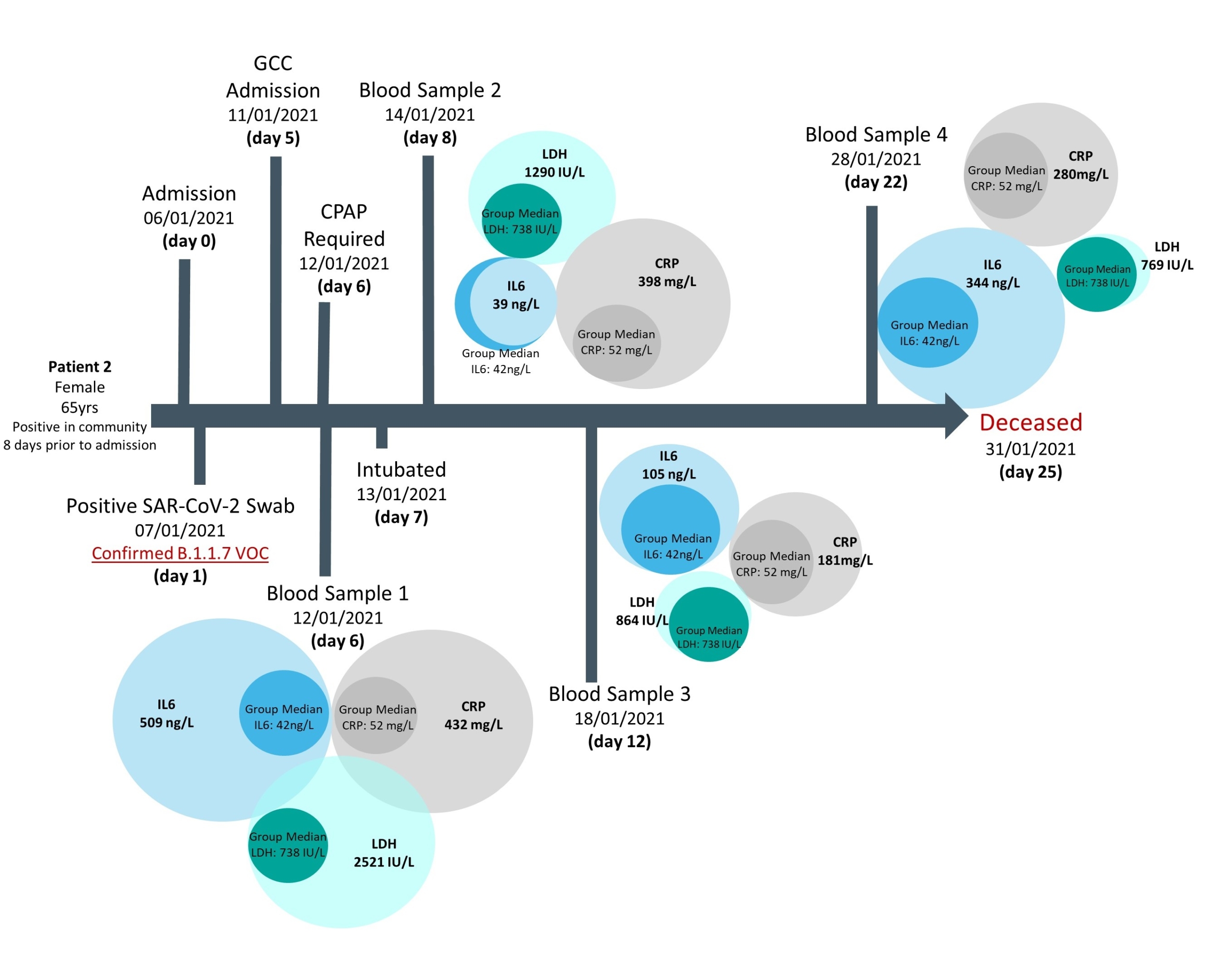
Access to a small cohort of admitted patients with confirmed VOC status also afforded the opportunity to follow biomarker longitudinal changes and correlate these according to disease outcomes, in particular mortality at the end of hospitalization. Results showed that in general across all cases investigated, deceased patients had raised values for several biomarkers in
comparison to the overall group median, whereas survivors had values closer to the median, supporting previously published findings (Figs 3 & 4). However, this approach in following biomarker trajectories provides additional information; allowing comparison of biomarker relationships across both the course of disease and in relation to one another. These patient pathways also highlight the fluctuation in biomarker values seen across the course of the disease and may provide guidance as to optimal timing for blood sampling or the need for multiple samples, when trying to develop predictive models for severe outcomes.
Summary
For poorly characterized diseases such as COVID-19, it is conceivable that many biomarkers have multiple pathophysiological determinants and can fluctuate for a wide range of mechanistic reasons. Therefore, biomarkers rarely provide a direct window into the underlying pathological processes of interest, and we recognize that individual biomarkers do not necessarily change linearly with, or indepen-dently of, other aspects of pathophysiology. The value and relative changes can depend heavily on other biomarkers, disease state severity and complications, and other crucial determinants such as comorbidities and demographics. However, advancing our understanding on relationships between biomarkers and out-
comes, can refine decision tree models to have more predictive power and can guide more precise approaches to understand disease pathogenesis.
Figure 1. Daily number of COVID-19 patients admitted to UK hospitals as reported by the UK Government from the start of the pandemic
(Data sourced from Coronavirus (COVID-19) in the UK, UK Government website [5])
Figure 2. Box plots highlighting mean and range of logarithmically transformed biomarkers from the COVID panel across two time periods
Figure 3. Biomarker levels during hospitalization of a confirmed B.1.1.7 infection case, where patient survival was the outcome of admission
Median biomarker values of patients with confirmed Alpha infections are provided as comparison.
Figure 4. Biomarker levels during hospitalization of a confirmed B.1.1.7 infection case, where patient death was the outcome of admission
Median biomarker values of patients with confirmed Alpha infections are provided as comparison.
The authors
Emma Braybrook1,2,3 MSc, Neil Anderson1,3 PhD and Dimitris Grammatopoulos*1,2,3 PhD
1 Institute of Precision Diagnostics and Translational Medicine, Pathology, UHCW NHS Trust, Coventry, UK
2 Translational Medicine, Warwick Medical School and Health Global Research Priority, University of Warwick, Coventry, UK
3 Department of Biochemistry and Immunology, Coventry and Warwickshire pathology Services, UHCW NHS Trust, Coventry, UK
*Corresponding author
E-mail: D.Grammatopoulos@warwick.ac.uk
References
1. Huang C, Wang Y, Li X et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395(10223):497–506 (https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(20)30183-5/fulltext).
2. Guidance on the use and interpretation of clinical biochemistry tests in patients with COVID-19 infection. Royal College of Pathologists 2020 (https://www.rcpath.org/uploads/assets/3f1048e5-22ea-4bda-953af20671771524/G217-RCPath-guidance-on-use-and-interpretation-of-clinical-biochemistry-tests-in-patients-with-COVID-19-infection.pdf).
3. Lauring AS, Hodcroft EB. Genetic variants of SARS-CoV-2 – what do they mean? JAMA 2021;325:529–531 (https://jamanetwork.com/journals/jama/fullarticle/2775006).
4. Tracking SARS-CoV-2 variants. World Health Organization 2022 (https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/).
5. Coronavirus (COVID-19) in the UK. UK Government 2022
(https://coronavirus.data.gov.uk/details/healthcare).
6. Murgolo N, Therien AG, Howell B et al. SARS-CoV-2 tropism, entry, replication, and propagation: Considerations for drug discovery and development. PLoS Pathog 2021;17(2):e1009225 (https://doi.org/10.1371/journal.ppat.1009225).
7. Gao W, Lv J, Pang Y, Li L. Role of asymptomatic and pre-symptomatic infections in covid-19 pandemic. BMJ 2021;375:n2342 (https://www.bmj.com/content/375/bmj.n2342.long).
8. Ning Q, Wu D, Wang X et al. The mechanism underlying extrapulmonary complications of the coronavirus disease 2019 and its therapeutic implication. Sig Transduct Target Ther 2022;7(1):57 (https://doi.org/10.1038/s41392-022-00907-1).
9. Malik P, Patel U, Mehta D et al. Biomarkers and outcomes of COVID-19 hospitalisations: systematic review and meta-analysis. BMJ Evid Based Med 2021;26(3):107–108 (https://ebm.bmj.com/content/26/3/107.long).
10. Davies NG, Abbott S, Barnard RC et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 2021;
372(6538):eabg3055 (https://doi.org/10.1126/science.abg3055).
11. Lin L, Liu Y, Tang X, He D. The disease severity and clinical outcomes of the SARS-CoV-2 variants of concern. Front Public Health 2021;9:775224 (https://doi.org/10.3389/fpubh.2021.775224).
12. Braybrook E, Pandey S, Vryonis E et al. Screening for the alpha variant of SARS-CoV-2 (B.1.1.7) the impact of this variant on circulating biomarkers in hospitalised patients. medRxiv 2021 (https://doi.org/10.1101/2021.06.18.21258699).