Getting the most from every drop – new advances in blood separation
Getting the most from every drop – new advances in blood separation
Alexandra Sommer, Senior Product Manager, Clinical Diagnostics at Tecan
Liquid biopsies are poised to play a central role in the future of diagnostics, especially in oncology.1 Currently, a conventional tissue biopsy is the gold standard in the diagnosis of many malignancies, but this technique comes with many drawbacks, including its invasive nature, the risk to patients, the need to fix the samples, and the difficulty in applying it to all cancer types and patients. Liquid biopsy allows genomic and proteomic assessment of components in the blood, or other sampled fluids, as an alternative, less invasive and more convenient method of monitoring cancer patients during and after treatment. Following sampling, the buffy coat and plasma can be separated from the erythrocytes by centrifugation, and analysed to understand the health status of the patient, and also check sample quality. Individual biomarkers in both the plasma and buffy coat – including circulating tumour cells (CTCs), circulating tumour DNA and circulating tumour ribonucleic acid (ctDNA, ctRNA), and exosomes – can be analysed to provide further insights into a patient’s cancer, offering a comprehensive view of tumour heterogeneity, and giving an overview of tumour characteristics such as progression, staging, heterogeneity, gene mutations, and clonal evolution. This can assist clinicians with devising personalised therapeutic regimens, as well as allowing continuous monitoring – by repeated sampling – to measure treatment efficacy and screen for therapeutic resistance.
CTCs
CTCs are intact tumour cells that are released into the bloodstream from primary or secondary tumour sites, and their migration through the circulatory system results in the development of distant metastases. They can be collected in the buffy coat following blood separation by centrifugation, and analysis of CTCs provides diagnostic and prognostic information in a number of cancer types. Their detection and characterisation is an important tool for monitoring and preventing the development of overt metastatic disease, and CTCs have also shown potential as screening biomarkers for early detection of cancers.2
ctDNA
Cancer cells shed naked DNA molecules, known as ctDNA, into the bloodstream. This genetic material can be isolated from plasma, and has become one of the main markers to investigate the mutational status of cancers through the use of next generation sequencing. Comparison of ctDNA with cfDNA – a normal product of cell turnover that is detectable in healthy individuals – can be used to identify mutations in tumours are not an exact match to an individual’s DNA, providing highly specific markers for various cancers.3 Recent advances in precision medicine have also allowed analysis of ctDNA to guide treatment decisions, predicting response and resistance to targeted therapies and immunotherapies. While most current clinical practice using ctDNA has been aimed at identifying druggable and resistance mutations for directing systemic therapy decisions, newer research is evaluating its potential as a marker of minimal residual disease, and as a useful screening tool to detect cancers in the general population.4
Exosomes
Exosomes are nano-vesicles released by various cell types, and carry a variety of ‘molecular messengers’ – including proteins, RNAs, DNAs and lipids. There is substantial evidence that exosomes are involved in intercellular communication by exchanging these messenger molecules between cells, and that they play important roles in cancer development. They can be isolated from plasma following blood separation, and recent studies have shown that exosomes are superior to CTCs and ctDNA for early diagnosis, disease monitoring, and prognostic prediction. However, they are more challenging to extract and study than other blood constituents, so improved strategies to isolate exosomes from bodily fluids – and to profile exosomal contents in a fast and sensitive way – are required to allow the practical application of exosome-based liquid biopsies for precision cancer medicine.5
Liquid biopsy applications also go beyond cancer. For example, non-invasive prenatal testing (NIPT) is becoming a preferred choice for many mothers undergoing testing for chromosomal abnormalities, as a safer alternative to amniocentesis. This is an increasingly important consideration as the average age of mothers continues to increase,6 prompting significant research interest in this area. Liquid biopsy approaches are also showing promise for monitoring patients following organ transplant. For example, a number of biomarkers – including donor-derived cell-free DNA, microRNAs and exosomes – are showing high potential for determining the long-term prognosis for kidney transplantation patients.7
The challenge of separating blood samples
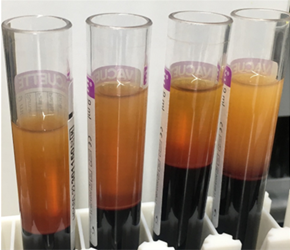
Blood collected for liquid biopsy must be separated into its constituents – plasma, buffy coat and erythrocytes – in order to study the markers described above. Traditional manual separation techniques are slow, and are susceptible to human error, even when performed by an experienced technician. Following centrifugation, the delicate buffy coat forms a thin layer between the plasma and erythrocytes, and manual isolation – usually carried out with a Pasteur pipette – carries the risk of contamination between the phases. Even when automation technology is available, it can be challenging to identify liquid-liquid interfaces, because barcodes and labels may interfere with optical sensors. With the increasing number of applications for liquid biopsies – from cancer monitoring to NIPT – laboratories need ways to streamline the workflow and overcome the bottleneck of blood separation.
(A) centrifuged blood
(B) after plasma separation
(C) after buffy separation