Mass spectrometric immunoassay for top-down protein analysis
Mass spectrometry-based methods hold great promise for addressing protein heterogeneity. As a result of post-translational processing, proteins can exist in vivo as multiple proteoforms. The added information contained in the protein profile can be important in physiological and pathological states. Presented here is an overview of a mass spectrometric immunoassay (MSIA) for quantitative determination of the chemokine RANTES proteoforms. MSIA offers protein quantification and profiling in a high-throughput and time-efficient manner. Across a cohort of ~300 human plasma samples, a total of 11 different RANTES proteoforms were quantified in less than 3 hours.
by Dr O. Trenchevska, N. D. Sherma, Dr P. D. Reaven, Dr R. W. Nelson and Dr D. Nedelkov
The role of mass spectrometry in protein analyses
Mass spectrometry (MS) has proven successful in the clinical laboratory for the analysis of small molecules, but is on the rise as an emerging methodology for peptides and proteins [1]. Currently, a handful of MS-based protein assays have been adapted in the routine clinical analyses and used for in vitro diagnostic (IVD) testing [2, 3]. MS-based methodologies are the assays of choice because they can overcome the limitations of immunoassays (i.e. nonspecific binding, cross-reactivity of analytes, etc.). In order to be clinically applicable, all MS-based assays should comply with the well-established ‘fit-for-purpose’ approach and be fully validated and characterized [4]. Also, working protocols must be practical (in terms of sample preparation), as well as cost efficient, so they are price-competitive with current immunoassays. Although overcoming these requirements is still a challenge, one inevitable advantage that makes MS-based protein assays indispensable, is their unique ability to address protein heterogeneity.
The majority of clinically adapted MS-based methodologies for protein profiling are the single/multiple reaction monitoring liquid chromatography MS (SRM/MRM LC-MS) assays [5, 6] and mass spectrometric immunoassays (MSIA) [7, 8]. MRM assays are ‘bottom-up’ assays and use isotopically labelled peptides as internal reference standards for surrogate protein quantification via chosen, enzymatically generated peptides. Because SRM/MRM LC-MS assays detect only specific peptides, important information about novel proteoforms or post-translational modifications with potential clinical implications can be overlooked. MSIAs, on the other hand, follow a ‘top-down’ approach, having intact proteins as primary targets. As a result of the immunoaffinity capture of a targeted protein(s), and the ‘soft’ ionization in MALDI-TOF (matrix-assisted laser desorption/ionization–time of flight) MS, MSIA enable for detection of post-translationally modified proteoforms as well as other changes in protein structure without the harsh enzyme digestion. Literature data show that post-translationally modified proteins have the potential to be used as biomarkers [9]. Having that in mind, the proteoform detection adds a whole new dimension to the way we look at proteins.
Mass spectrometric immuno-assay for analysis of RANTES proteoforms
Here we review a mass spectrometric immunoassay (MSIA) for quantification of the chemokine RANTES proteoforms in human plasma samples. RANTES (Regulated on Activation, Normal, T-cell Expressed and Secreted), is a member of the CC chemokine family (hence its alternative name – CCL5) and is essential in the initiation and maintenance of inflammation [10]. RANTES has been studied extensively in clinical context, in association with autoimmune diseases, arthritis, diabetes, obesity and metabolic syndrome, some types of cancer and viral infections [11–13]. In addition, RANTES proteoforms have been associated with atherosclerosis and cardiovascular diseases [14].
There are several types of commercially available, as well as in-house developed immunoassays for total RANTES quantification [15]. These assays, however, are not tailored for detecting and quantifying the numerous proteoforms associated with RANTES. In previous work, we have addressed RANTES heterogeneity by qualitative and quantitative MSIA [16, 17]. In developing the quantitative MSIA for RANTES, we took on the approach of using RANTES standard and a homologous RANTES derivative – met-RANTES as an internal reference standard (IRS) for quantification. Met-RANTES is a recombinant derivative of RANTES (therefore not found in humans) and has a molecular weight (MW) of 7979.2 Da, which is in close proximity to that of full-length human RANTES (MW=7847.9 Da). Another advantage of using the RANTES/met-RANTES pair was the ability of a single anti-RANTES antibody to capture both proteins from the biological samples.
The immobilization of the anti-RANTES antibody was onto activated surfaces of affinity pipettes as previously described [17]. The quantity of the anti-RANTES antibody (7.5 µg Ab/tip) was optimized to be enough that variable RANTES concentrations in the samples could be truly quantified with the assay. Due to low plasma RANTES physiological concentration (in the ng/mL level), undiluted plasma was used for the analyses. In the analytical samples, met-RANTES was spiked at a constant concentration (V=250 µL at c=50 ng/mL), in order to produce a constant signal in the mass spectra. Following sample preparation and affinity pipette derivatization, the antibody-coated pipettes were mounted onto the head of an automated 96-channel pipettor and initially rinsed with PBS/0.1% Tween buffer. Next, the pipettes were immersed into a microplate containing the analytical samples and 500 aspirations and dispense cycles were performed (100 μl volumes each) allowing for affinity capture of RANTES proteoforms and met-RANTES. The pipettes were then rinsed with assay buffer water to remove non-specifically bounded proteins. Captured proteins were eluted directly on a 96-well formatted MALDI target using sinapic acid. Five-thousand laser shots of mass spectra were acquired from each sample spot on a Bruker’s Ultraflex III MALDI-TOF/TOF mass spectrometer. The mass spectra were externally and internally calibrated with protein standard mix and the singly and doubly charged met-RANTES signals before analysis.
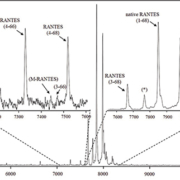
In the mass spectra, several RANTES proteoforms can be detected. As shown in Figure 1, most abundant are signals representing full-length, native RANTES (1-68) and met-RANTES, along with the N-terminally cleaved RANTES proteoforms (3-68) [MW=7,663.7; missing the ‘SP’ N-terminal dipeptide, product of dipeptidyl peptidase IV (DPP IV) enzyme cleavage] and (4-68) (MW=7,500.6; missing ‘SPY’ N-terminal tripeptide). RANTES proteoforms missing N-terminal tripeptide and C-terminal dipeptide, (4-66) (MW=7,282.3) completed the dominant signals (Figure 1, top right inlet). Additional RANTES proteoforms were identified, in lower abundance and frequency: (7-66) (MW=6993.1; missing six N-terminal and two C-terminal amino acids), (4-64) (MW=7040.1; missing three N-terminal and four C-terminal amino acids), (4-65) (MW=7153.2; missing three N- and three C-terminal amino acids) and (3-66) (MW=7445.5; missing two N- and two C-terminal amino acids). The signal labelled M-RANTES with MW=7413.5 has multiple N- and C-terminal truncation possibilities, and has not been specifically assigned. The assignation of these signals was done using the observed m/z values and the program Paws, and was in accordance with previously published qualitative results [16].
All identified RANTES proteoforms were quantified using an eight-point standard curve, in the range from 1.56 to 200 ng/mL. The standard curve was constructed from the ratio of the peak intensities of the RANTES standard and the met-RANTES IRS (y-axis) versus the RANTES standard concentration (x-axis). For the analytical samples, first, the RANTES/met-RANTES peak intensity ratios for each proteoform were determined and summed up. Using the generated standard curve equation, these ratios were used to determine the total RANTES concentration in the analysed plasma sample. Then, the concentration of the individual RANTES proteoforms was calculated based on their percentage of the total RANTES. The assay was validated through several standard procedures. The intra- and inter-assay precision experiments yielded coefficients of variation of <10%. Linearity and spiking-recovery experiments produced results between 92 and 112% (observed vs expected concentration). In a final test, the results of the RANTES MSIA were compared with those obtained with commercially available ELISA using Altman–Bland plot. A good correlation, with slight positive bias (11.3%) was obtained with the native RANTES [17].
The developed MSIA for RANTES proteoforms was applied to a cohort of 297 human plasma samples. The analyses were performed on an automated platform, which enabled for a high-throughput analysis of 96 samples in a single run. Among the samples, we were able to determine the concentration and frequency of 11 RANTES proteoforms (Figure 2). The total average concentration of RANTES was found to be 44.9 ng/ml (2.15–163 ng/mL). In majority of samples, the main proteoform was the full-length, native RANTES [c(RANTES(1-68))avg=37.4 ng/mL; 1.92–132 ng/mL], followed by RANTES (3-68), [c(RANTES(3-68))avg =6.64 ng/mL; 0.138–34.4 ng/mL]. The other truncated RANTES proteoforms were present in variable frequencies in the samples, albeit at much lower concentrations (<10% of the total RANTES). Figure 2 summarizes the distribution and frequency of all 11 RANTES proteoforms. Even though majority of RANTES proteoforms were detected in only a handful of samples and in low quantities, they should be given full attention. Cleaved proteoforms have the potential to be used as indicators of an enzymatic activity, and, in turn, of changes in the metabolic homeostasis [18]. The information that this MSIA provides puts a new perspective of RANTES quantitative analysis and can be a good starting point for looking at RANTES heterogeneity in clinical context.
Concluding remarks
The assay described above uses MALDI-TOF-MS to fully quantify RANTES proteoforms, and it is one of just a handful of such MALDI-based assays in existence today. The assay’s two-step approach is similar to that of well-established immunoassays, with the added benefit of MS detection as an enabling factor in differentiating the multiple proteoforms. The MALDI target is designed to accept the eluates from 96 tips at the same time, therefore making it high-throughput and time efficient (total time for RANTES assay is ~1 hour). The assay is performed on an automated platform, which limits the errors that can occur during assay execution. In review of previous and ongoing work, MSIA for RANTES performs well and introduces a new prospect and capacity for potential clinical applications in the field of biomarker discovery/rediscovery and diagnostics.
References
1. Strathmann FG, Hoofnagle AN. Am J Clin Pathol. 2011; 136: 609–616.
2. Agger SA, Marney LC, Hoofnagle AN. Clin Chem. 2010; 56: 1804–1813.
3. Kiernan UA, Phillips DA, Trenchevska et al. PLoS One 2011; 6: e17282.
4. Carr SA, Anderson L. Clin Chem. 2008; 54: 1749–1752.
5. Anderson NL, Anderson NG, Haines LR, et al. J Proteome Res. 2004; 3: 235–244.
6. Yocum AK, Chinnaiyan AM. Brief Funct Genomic Proteomic. 2009; 8: 145–157.
7. Nelson RW, Krone JR, Bieber AL, et al. Anal Chem. 1995; 67: 1153–1158.
8. Trenchevska O, Kamcheva E, Nedelkov D. Proteomics 2011; 11: 3633–3641.
9. Jin H, Zangar RC. Biomark Insights 2009; 4: 191–200.
10. Youn BS, Mantel C, Broxmeyer HE. Immunol Rev. 2000; 177: 150–174.
11. Lit LC, Wong CK, Tam LS, et al. Ann Rheum Dis. 2006; 65: 209–215.
12. Matter CM, Handschin C. Circulation 2007; 115: 946–948.
13. Azenshtein E, Luboshits G, Shina S, et al. Cancer Res. 2002; 62: 1093–1102.
14. Winnik S, Klingenberg R, Matter CM. Eur Heart J. 2011; 32: 393–395.
15. Kaburagi Y, Shimada Y, Nagaoka T, et al. Arch Dermatol Res. 2001; 293: 350–355.
16. Oran PE, Sherma ND, Borges CR, et al. Clin Chem. 2010; 56: 1432–1441.
17. Trenchevska O, Sherma ND, et al. J Proteomics 2014; 116C, 15–23.
18. Lim JK, Lu W, Hartley O, et al. J Leukoc Biol. 2006; 80: 1395–1404.
The authors
Olgica Trenchevska*1, Nisha D. Sherma1, Peter D. Reaven2, Randall W. Nelson1, Dobrin Nedelkov1
1Molecular Biomarkers, The Biodesign Institute at Arizona State University, Tempe, AZ, USA
2Phoenix Veterans Affairs Health Care System, Phoenix, AZ, USA
*Corresponding author
E-mail:
olgica.trenchevska@asu.edu