Molecular diagnosis for eosinophilic esophagitis: a next-generation technique with pathogenic insight
Eosinophilic esophagitis (EoE) is a clinical disorder induced by food allergy with its current diagnosis based on histological examination of esophageal biopsies and clinical symptoms. A next-generation molecular diagnostic panel based on a 96-gene qPCR array was recently introduced for a definitive and objective diagnosis of EoE and demonstrated high diagnostic merit relative to the current method. This test provides insight into the pathological processes of EoE in a cost effective manner and will likely bring a personalized medicine approach to the field of esophagitis.
by Dr Ting Wen and Professor Marc E. Rothenberg
Eosinophilic esophagitis and current form of diagnosis
The tide of technical advances in the new century has sparked a molecular revolution in the fields of clinical diagnostics and predictive medicine, and is crucial for the provision of personalized medicine. Thus far, molecular diagnosis of diseases has been largely confined to cancer, autoimmunity and genetic disorders, while areas such as gastrointestinal (GI) disorders and allergic inflammation have been underexplored.
Eosinophilic esophagitis (EoE) is a type of immunological food allergy mediated by allergic hyper-responses to food, typically the six most common food allergens (milk, egg, wheat, soy, nuts, fish). Dietary elimination and steroidal intervention are two of the most effective therapies, and are often used together. At the cellular level, the inflammation is a well concerted process caused by local lymphocytes (primarily Th2 cells), mast cells and eosinophils within the esophagus that likely contributes to most of the clinical symptoms. Clinically, EoE is characterized by esophageal dysfunction (e.g. dysphagia) and is historically defined by a tissue biopsy exhibiting ≥15 eosinophils per high-power field (EOS/HPF), a cut-off agreed by a panel of field experts based on case discussions 5 years ago and referred to as the consensus recommendation (CR) 2007 [1, 2]. Both histological and clinical symptoms are necessary for diagnosis [ideally with proton pump inhibitor (PPI) trial confirmation] and form the basis of CR2007. Thus far, histological examination is the only widely accepted EoE diagnostic test [2]. However, this method is subjective, time-consuming and subject to variability (patchy sampling and inter-pathologist variability), as well as expensive. In addition, it is non-specific to a certain extent, as there are a number of other diseases sharing esophageal eosinophilia [3], and therefore the histological method cannot identify specific exposure to medications (such as glucocorticoid) nor differentiate patients in remission from EoE from non-EoE patients (neither exhibit eosinophilia). Therefore, the current ‘gold standard’ method has limitations and its results may be questionable. [4].
EoE transcriptome, qPCR array and algorithm development
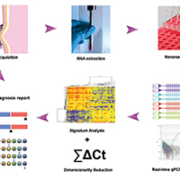
The EoE transcriptome was identified by Blanchard et al. in 2006 [5] and subsequent studies identified ~1000 genes that were bi-directionally dysregulated at different magnitudes; thus serving as the foundation for molecular differentiation. We adopted a low density array in which four identical custom arrays are embedded on a 384-well fluidic card (Fig. 1), which allows cost-efficient Taqman-based PCR to be performed on potentially informative genes that are part of the EoE transcriptome. With this design, a maximum of 95 genes (plus one housekeeping control) can be assayed to map molecular pathogenic signatures and four samples can be processed in a given batch thus improving the turn-around time. During a recent study that involved a large cohort of 194 unique samples and used the qPCR-based array described above (termed the EoE diagnostic panel, or EDP), the test performed at nearly 100% accuracy in terms of revealing the bi-directional EoE signature, at much lower cost and with faster turn-around (same-day) compared with the standard microscopic analysis [6]. Notably, the EDP is designed to work with both fresh tissue and formalin-fixed, paraffin-embedded (FFPE) samples, with major steps involving RNA isolation, reverse transcription, qPCR amplification, raw data rendering, a dual algorithm application and diagnostic report generation (Fig. 1) (Table 1).
The test is highly reproducible between batches and samples, as results from samples tested months apart still correlated well because of the accuracy and specificity of Taqman qPCR. Having generated the raw cycle threshold (Ct) values of qPCR, the critical next step was to develop a method of interpreting the results in a way that every physician and patient was able to comprehend. One of the unique features of the EDP is the novel dual algorithm associated with the panel, which provides additional assurance. Briefly, the first algorithm is a clustering analysis based on the Pearson correlation of 77 genes followed by dimensionality reduction. With 50 upregulated genes and 27 downregulated genes, the bi-directional dysregulation provides a pronounced contrast for signature recognition. A dendrogram (hierarchical tree) is then derived from the inter-sample distance metrics aided by commercial analysis softwares such as GeneSpring (Agilent Inc.). The first branch of the dendrogram serves as a diagnostic bifurcation point. The second algorithm performed in parallel is essentially a mathematical summing-up of the change in Ct value for each gene relative to the housekeeping gene (GADPH) and takes into account the bi-directional changes (+ and – vectors). The end read-out of this algorithm is an absolute integer that provides a definitive EoE diagnosis and which correlates linearly with disease severity. This direct output allows the physician to readily assess the disease status and potential therapy. These types of dual algorithms are not mathematically challenging, so it is expected that the same algorithm may be expanded for broader use in the diagnosis of other inflammatory diseases, especially those with a diagnostic ‘grey zone’ or other clinical dilemma.
Data management, FFPE compatibility and commercialization potential
Compared to other more advanced gene-expression platforms, the Taqman qPCR array-based EDP has the advantage of cost effectiveness, minimal data size and straightforward experimental analysis. Basically, the raw Ct data are exported from the qPCR machine (ABI 7900HT) in text (.txt) or Excel format (typically in seconds), and are then further processed by software such as GeneSpring or simply by formula calculation in Excel. The goal is to generate an ‘EoE score’ and a clustered heat diagram leading to definitive diagnosis and disease evaluation on the basis of patient specific profiles of the 95-gene signature, therefore enabling a form of personalized medicine.
Of note, the EDP offers good compatibility with FFPE samples, as shown by a sub-study with 45 randomly selected FFPE samples, reaching 96% sensitivity and 100% specificity using histology as the ‘gold standard’. While FFPE RNA is known to be susceptible to considerable degradation with time (also shown in this study by Agilent QC assay), the EDP signature acquisition does not seem to be compromised over the reported archiving duration of 3 years. One can only imagine how many clinical questions could be answered using vast archived pools of FFPE samples and the associated amount of clinical outcome information.
With the simple qPCR-based procedures and the easily accessible computational algorithm, as well as the demonstrated clinical utility, the commercialization potential of the EDP is promising. The future use of the EDP in clinical practice is not reliant on complicated techniques, advanced hardware, professional expertise or significant start-up funds. Together with the rapid turn-around time and minimal demands on lab personnel, the initial barrier for adoption of the EDP in practice is low. It is also worth mentioning that such novel qPCR arrays combined with novel scoring algorithms could be readily applied to the diagnosis of other allergic inflammatory diseases with minimal modifications.
Conclusions and future expectations
Given the growing interest in “next-gen” molecular techniques, there seems to be ample justification for the development of this novel diagnostic method. Compared to the classic histology-based EoE diagnosis, there are several aspects that the molecular method is uniquely capable of providing. First, the molecular panel is able to distinguish remission patients despite normal histology, which is clinically important and not achievable using standard histology. The EDP is essentially based on reading gene expression in 96 channels (96-D), providing multi-dimensional pathogenic clues and personalized medical information compared with histology and other reported immunohistochemistry (IHC)-based approaches. Recently, a 4-protein panel was reported as having high diagnostic merit [7]. Although having a greater variety of diagnostic methods for EoE is beneficial, the lengthy IHC workload, subjective interpretation and insufficient pathogenic resolution make the molecular method advantageous. Preliminary results also suggest that the EDP can be used to predict the likelihood of EoE relapse prospectively, indicating a predictive medicine component of this technique. The EDP also provides a means to assess steroid exposure based on steroid responding gene elements, which could be used to evaluate patient compliance. Finally, regarding sensitivity and specificity, there is currently no option but to continue to use histology as the ‘gold standard’, because it is not yet clear which method more objectively reflects the disease status. Further research is needed to clarify this, especially in the light of the debatable specificity of histology [3, 4]. Although the EDP doesn’t provide all the answers, for example there is no predictive component for identifying which type of treatment will be more successful (diet versus steroid) and the sample source is still deemed invasive, it nevertheless represents a substantial advance. The conventional method has its merits, namely directly visualizing infiltrating eosinophils (for which EoE was named) and cellular topology, and will continue to be used in parallel. However, the advantages of the new platform are likely to be appreciated by researchers and physicians in an era where disease definition and pathogenic understanding are increasingly at the molecular level.
References
1. Furuta GT, Liacouras CA, Collins MH, Gupta SK, et al. Eosinophilic esophagitis in children and adults: a systematic review and consensus recommendations for diagnosis and treatment. Gastroenterology 2007; 133(4): 1342–1363.
2. Liacouras CA, Furuta GT, Hirano I, Atkins D, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011; 128(1): 3–20 e6; quiz 21–22.
3. Dellon ES, Gonsalves N, Hirano I, Furuta GT, et al. ACG clinical guideline: Evidenced based approach to the diagnosis and management of esophageal eosinophilia and eosinophilic esophagitis (EoE). Am J Gastroenterol. 2013; 108(5): 679–692; quiz 693.
4. Rodrigo S, Abboud G, Oh D, DeMeester SR, et al. High intraepithelial eosinophil counts in esophageal squamous epithelium are not specific for eosinophilic esophagitis in adults. Am J Gastroenterol. 2008; 103(2): 435–442.
5. Blanchard C, Wang N, Stringer KF, Mishra A, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006 116(2): 536–547.
6. Wen T, Stucke EM, Grotjan TM, Kemme KA, et al. Molecular diagnosis of eosinophilic esophagitis by gene expression profiling. Gastroenterology 2013; doi:10.1053/j.gastro.2013.08.046.
7. Dellon ES, Chen X, Miller CR, Woosley JT, Shaheen NJ. Diagnostic utility of major basic protein, eotaxin-3, and leukotriene enzyme staining in eosinophilic esophagitis. Am J Gastroenterol. 2012 107(10): 1503–1511.
The authors
Ting Wen PhD and Marc Rothenberg* MD, PhD
Division of Allergy and Immunology, Cincinnati Children’s Hospital Medical Center, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA
*Corresponding author
E-mail: Rothenberg@cchmc.org