Molecular diagnostics from stained cytology smears
Molecular testing is increasingly recognized for guiding patient management and development of new targeted therapies. Meanwhile, there is an increased demand to perform testing on smaller volumes of tissue. Recent literature reports the use of ultrasensitive techniques to detect DNA mutations and translocations from stained cytology smears [1].
by Dr Laleh Hakima, Dr Maja H. Oktay and Prof. Sumanta Goswami
Introduction
Mutational analyses are crucial for guiding treatment decisions. This is particularly important for targeted therapies with tyrosine kinase inhibitors (TKI) for lung non-small cell carcinoma (NSCC) and for surgical management of thyroid nodules with indeterminate cytological diagnoses. Both lung and thyroid lesions are frequently diagnosed using fine need aspiration (FNA). FNA is a preferred method of obtaining diagnostic samples compared to surgical excisions or core biopsies. The FNA procedure is minimally invasive, rapid, cost-effective and has reduced procedure-related complications.
Molecular diagnostic tests of cytological samples are often performed using cytology cell block preparations. However, insufficient cellularity of cell blocks is a frequent limitation to these tests. In addition, errors introduced by formalin fixation may also interfere with accurate detection of mutations. Recent studies have reported successful testing using cytology smears stained with Diff-Quik®, which are air dried and fixed in alcohol. This approach has several advantages compared to testing on paraffin-embedded tissue (PET). Alcohol is a great preservative for DNA and RNA. The sample quality is superior to PET because FNA samples are typically enriched for cancer cells and cytology smears yield intact whole nuclei rather than the nuclear fragments that are obtained by cutting PET [2]. Lastly, testing performed directly on microdissected tumour cells ensures that material is sufficient and representative of the tumour and results in a faster turnaround time. We used cytological direct smears stained with Diff-Quik® and Papanicolaou (Pap) to detect commonly encountered mutations in non-small cell lung carcinoma and thyroid carcinoma [1].
Lung- and thyroid-carcinoma mutations and fusion gene testing targets
Approximately 64% of all lung adenocarcinomas harbour somatic driver mutations. According to The Lung Cancer Mutation Consortium, the frequency of EGFR and KRAS mutations are 23% and 25%, respectively. The incidence of EML4–ALK translocation, mainly detected in non-smoker patients with wild-type EGFR and KRAS genes, is approximately 6% [2, 3]. Other less frequent mutations include: BRAF ~3%, PIK3CA ~3%, MET amplifications ~2%, ERBB2 (HER2/NEU) ~1%, MAP2K1 ~0.4%, and NRAS ~0.2% [2]. Additionally, new driver mutations have recently been identified in lung cancer patients [3]. The most common mutations in thyroid carcinoma involve BRAF, KRAS, and RET/PTC genes. The mutations in BRAF and KRAS are point mutations, whereas RET (PTC) mutations are gene rearrangements that result in fusion of the tyrosine kinase domain of the RET gene to various unrelated genes [4–7].
The EGFR mutation status of the cancer is associated with its responsiveness or resistance to EGFR TKI therapy. The EGFR gene is located on chromosome 7p11.2, spans about 200kb, and contains 28 exons. The gene encodes a protein of 464 amino acids. The EGFR protein is composed of an N-terminal extracellular ligand-binding domain, a transmembrane lipophilic segment, and a C-terminal intracellular region containing a tyrosine kinase domain. The EGFR tyrosine kinase modulates cell proliferation and survival. Activation of EGFR initiates signalling cascades involving several downstream pathways, including Ras GTPases, which induce crucial cellular responses, such as proliferation, differentiation, motility, and survival. EGFR mutations associated with objective responses to single-agent TKI therapy in lung adenocarcinomas are preferentially observed in females of East Asian ethnicity who are never smokers and have adenocarcinoma with lepidic growth pattern (formerly bronchioloalveolar carcinoma). In adenocarcinomas, the majority of mutations have been identified in exons 18–21 of the EGFR gene. These mutations can be roughly classified into three major categories: in-frame deletions in exon 19, insertion mutations in exon 20, and missense mutations in exons 18–21 [8].
The fusion of the echinoderm microtubule-associated protein-like 4 (EML4) gene to the anaplastic lymphoma kinase (ALK) gene, EML4–ALK, is the most common fusion and results from the joining of exons 1–13 of EML4 to exons 20–29 of ALK. At least seven EML4–ALK variants (V1–V7) have been identified in lung adenocarcinomas. All seven variants are formed through the fusion of the intracellular tyrosine kinase domain of ALK with a variably truncated EML4 gene promoter. Activated ALK is involved in the inhibition of apoptosis and the promotion of cellular proliferation through activation of downstream PI3K/AKT1- and MAPK1-signalling pathways. The key downstream effectors on the ALK pathway include the Ras-activated protein, mitogen-activated protein kinase 1 [MAPK1; also known as extracellular signal regulated kinase (ERK)], phosphatidylinositol 3-kinase (PI3K), and signal transducer and activator of transcription 3 (STAT3) signalling pathways. Ras/MAPKK1/MAPK1 pathways are critical for cell proliferation, whereas the PI3K/AKT1 and STAT3 pathways are important for cell survival. The histology of these tumours is typically characterized by mucin production and either a solid growth pattern containing signet ring cells in Western patients or an acinar growth pattern in Asian patients. Compared with patients with wild-type ALK and EGFR, patients with the EML4–ALK fusion gene tend to be younger, of Asian ethnicity, diagnosed at an advanced clinical stage at presentation, male dominant, and more likely to be never smokers, but with a comparable response rate to chemotherapy and overall survival. The EML4–ALK fusion gene is typically detected by fluorescence in-situ hybridization (FISH). It has been reported that although ALK-fusion-positive lung cancers are resistant to the EGFR TKIs, gefitinib, and erlotinib, they are sensitive to small molecule TKIs against ALK [8].
The BRAFV600E point mutation involves nucleotide 1799 and results in a valine-to-glutamate substitution at residue 600 (V600E). B-Raf is a serine/theronine protein kinase involved in MAPK/ ERK signalling pathway and in regulation of cell proliferation and differentiation. It is found in approximately 40–45% of papillary thyroid carcinomas. Not all variants are equally affected; 60% of classic papillary, 80% of tall cell variant, and 10% of follicular variant harbour this mutation. Its detection is clinically significant because if represents a prognostic marker for thyroid papillary carcinoma, it is associated with extrathyroidal extension, advanced tumour stage at presentation, and lymph node or distant metastases. BRAFV600E point mutation is also an independent predicator of treatment failure and tumour recurrence, even with patients with low-stage disease [4].
Case selection and molecular analysis techniques
Thirty-one cases of lung adenocarcinomas and 26 thyroid carcinomas (17 classic papillary, 7 follicular variant, and 2 follicular carcinomas) were selected from the archives. Molecular analysis was performed on PET and from either Pap or Diff-Quik® stained smears for each case. The following mutations in lung adenocarcinomas were tested: EGFR point mutations in exons 20 and 21, in-frame deletions in exon 19; KRAS point mutations in codons 12, 13, and 61; and EML4–ALK translocation. Thyroid carcinomas were tested for the BRAFV600E point mutation.
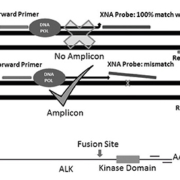
Smears were reviewed by two cytopathologists and areas containing at least 50 cancer cells without necrosis or inflammation were marked for analysis [9, 10]. Tumour cells from marked areas were microdissected using RNA/DNA co-purification solution (Zymo Technologies) [5, 6]. QClamp xenonucleic acid (XNA) technology was used to detect mutations in EGFR, KRAS and BRAF genes. The qClamp, a wild-type sequence-specific XNA probe, has a melting point higher than 72 °C and remains attached to the wild-type DNA during the PCR extension stage. If there is a mutation, the probe dissociates from the template, which allows amplification. The technology was optimized using wild-type DNA (Promega, Madison, WI) resulting in a DNA shift of more than seven cycle thresholds (CTs) with the XNA clamp. In samples with mutations, the shift was less than 5 CTs (Fig. 1).
Quantitative RT-PCR (RT-qPCR) was used to detect EML–ALK translocations. PCR primers were used to amplify both the 3ʹ and 5ʹ ends of the ALK transcript (Aanera Biotech). In the presence of the translocation, the transcription of the EML4–ALK fusion gene (under the control of a stronger EML promoter) resulted in higher than expected 3ʹ ends than 5ʹ ends which lead to lower CT values for the 3ʹ transcript. A difference of more than 10 CTs between 3ʹ and 5ʹ ends was considered positive (Fig. 1).
The sensitivity of our assay was determined using two lung cancer cell lines, H1975 and H2228 (ATCC), harbouring the L858R mutation in the EGFR gene and the EML4–ALK translocation, respectively. The mutation detection rate was 1%.
Results
Approximately 80.6% of lung cases had some form of molecular alteration detected. There was 100% concordance between PET and cytology smears. All cases tested positive by our laboratory were also positive by the reference laboratory. However, seven cases that tested negative for EGFR mutation by the reference laboratory were found to be positive in our lab. In other words, clamp qPCR methodology detected approximately 20% more EGFR mutations than the reference laboratory. Likewise, three cases that were negative for KRAS by the reference laboratory, tested positive in our laboratory. In addition, there were six cases (19%) that had more than one molecular alteration in our cohort. Five cases had two mutations (three had EGFR exon 19 and KRAS codon 12 mutations, two had KRAS codon 12 and EML4–ALK mutations) and one case had three mutations (EGFR exon 19 and 21 and KRAS codon 12 mutation). Although EGFR and KRAS mutations were previously thought to be mutually exclusive, our results confirm recent reports of simultaneous mutations detected in study samples [7]. Given the known heterogeneity of cancers this finding may be expected, but might not have been detected previously because most analyses are performed using standard-sensitivity techniques.
Of 26 thyroid cases, 18 (81%) were positive for BRAFV600E mutation on both PET and cytology smears. Concordant results were obtained from both cytology smears and PET from cases tested by us and the reference laboratory. Both follicular carcinoma cases tested negative on cytology smears and PET for BRAFV600E. The percentage of patients with a BRAF mutation detected by us was higher than expected from the literature (81% versus 50%) reflecting our ultrasensitive approach to mutation detection [10].
Conclusion
Our results indicate that qClamp technology and the RT-qPCR approach can be used successfully to detect most common targetable molecular alterations from cytology smears of lung and thyroid carcinomas. We report successful DNA and RNA isolation from both Diff-Quik® and Pap stained smears. This methodology represents an ultrasensitive and accurate approach with a 1% mutation detection rate and a decreased turnaround time of 1 to 2 days. Such an ultrasensitive method for molecular testing is essential as smaller amounts of diagnostic material become available and targeted approaches will be aimed at both molecular alterations present in the majority of cells, and at those present in a minority of cells that potentially may represent a subpopulation susceptible to recurrence [11].
Abbreviations
Table 1 shows the gene symbols and the names and symbols of the proteins encoded by those genes.
References
1. Oktay MH, Adler E, Hakima L, Grunblatt E, Pieri E, Seymour A, Khader S, Cajigas A, Suhrland M, Goswami S. The application of molecular diagnostics to stained cytology smears. J Mol Diagn. 2016; 18(3): 407–415.
2. Tsao MS, Sakurada A, Cutz JC, Zhu CQ, Kamel-Reid S, Squire J, Lorimer I, Zhang T, Liu N, Daneshmand M, Marrano P, da Cunha Santos G, Lagarde A, Richardson F, Seymour L, Whitehead M, Ding K, Pater J, Shepherd FA. Erlotinib in lung cancer – molecular and clinical predictors of outcome. N Engl J Med. 2005; 353: 133–144.
3. Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, Shukla SA, Guo G, Brooks AN, Murray BA, Imielinski M, Hu X, Ling S, Akbani R, Rosenberg M, Cibulskis C, Ramachandran A, Collisson EA, Kwiatkowski. DJ, Lawrence MS, Weinstein JN, Verhaak RG, Wu CJ, Hammerman PS, Cherniack AD, Getz G, Cancer Genome Atlas Research Network, Artyomov MN, Schreiber R, Govindan R, Meyerson M. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016; 48(6): 607–616.
4. Nikiforov YE. Molecular diagnostics of thyroid tumors. Arch Pathol Lab Med. 2011; 135(5): 569–577.
5. Gupta N, Dasyam AK, Carty SE, Nikiforova MN, Ohori NP, Armstrong M, Yip L, LeBeau SO, McCoy KL, Coyne C, Stang MT, Johnson J, Ferris RL, Seethala R, Nikiforov YE, Hodak SP. RAS mutations in thyroid FNA specimens are highly predictive of predominantly low-risk follicular-pattern cancers. J Clin Endocrinol Metab. 2013; 98(5): 914–922.
6. Nikiforov YE, Ohori NP, Hodak SP, Carty SE, LeBeau SO, Ferris RL, Yip L, Seethala RR, Tublin ME, Stang MT, Coyne C, Johnson JT, Stewart AF, Nikiforova MN. Impact of mutational testing on the diagnosis and management of patients with cytologically indeterminate thyroid nodules: a prospective analysis of 1056 FNA samples. J Clin Endocrinol Metab. 2011; 96(11): 3390–3397.
7. Nikiforov YE, Yip L, Nikiforova MN. New strategies in diagnosing cancer in thyroid nodules: impact of molecular markers. Clin Cancer Res. 2013; 19(9):2283–2288.
8. Cheng L, Alexander RE, Maclennan GT, Cummings OW, Montironi R, Lopez-Beltran A, Cramer HM, Davidson DD, Zhang S. Molecular pathology of lung cancer: key to personalized medicine. Mod Pathol. 2012; 25: 347–369.
9. Marchetti A, Milella M, Felicioni L, Cappuzzo F, Irtelli L, Del Grammastro M, Sciarrotta M, Malatesta S, Nuzzo C, Finocchiaro G, Perrucci B, Carlone D, Gelibter AJ, Ceribelli A, Mezzetti A, Iacobelli S, Cognetti F, Buttitta F. Clinical implications of KRAS mutations in lung cancer patients treated with tyrosine kinase inhibitors: an important role for mutations in minor clones. Neoplasia. 2009; 11: 1084–1092.
10. Russo M, Malandrino P, Nicolosi ML, Manusia M, Marturano I, Trovato MA, Pellegriti G, Frasca F, Vigneri R. The BRAF (V600E) mutation influences the short and medium-term outcomes of classic papillary thyroid cancer, but is not an independent predictor of unfavorable outcome. Thyroid 2014; 24: 1267–1274.
11. Pon JR, Marra MA. Driver and passenger mutations in cancer. Annu Rev Pathol. 2015; 10: 25–50.
The authors
Laleh Hakima1 DO; Maja H. Oktay1 MD, PhD; Sumanta Goswami*2 PhD
1Department of Cytopathology, Montefiore Medical Center Bronx, NY, USA
2Department of Biology, Yeshiva University, New York, NY, USA
*Corresponding author
E-mail: Goswami@yu.edu