Molecular diagnostics of pathogens using next-generation sequencing
Introduction
The correct identification and characterization of pathogens is essential for the successful treatment of infections and safety of patients. However, not every pathogen can be successfully cultured and the available molecular tests, mainly focusing on specific pathogens, are inadequate to detect novel genetic features in emerging pathogens. Undetected pathogens can spread easily through a hospital, resulting in a possible outbreak and putting patients admitted to hospitals at a higher risk for infections.
In recent decades, molecular diagnostic tests have improved rapidly and their role in clinical microbiology laboratories became progressively more important [1]. The turnaround time from receiving a sample to the final diagnostic result has been drastically reduced. Molecular methods, such as real-time polymerase chain reaction (PCR), Sanger sequencing and next-generation sequencing (NGS), make it possible to detect non-culturable micro-organisms. Nevertheless, some of these technologies, such as real-time PCR, require knowledge of the genomes of the micro-organisms. In addition, bioinformatics expertise is often needed to interpret the results.
This paper addresses the use of Sanger sequencing and whole genome sequencing (WGS) in the clinical microbiology laboratory for the characterization of pathogens and outbreak management, as it is used in the University Medical Center Groningen (UMCG), one of the largest university hospitals in The Netherlands. The clinical microbiology laboratory at the UMCG receives around 5750 samples per year for detailed molecular analysis, of which approximately 1500 samples are analysed by NGS [2].
Sanger sequencing
Sanger sequencing is used to answer different molecular questions, such as the identification of bacteria and fungi in patient material or pure cultures, and the identification of mutations in specific genomic regions of interest in bacteria or viruses. In general, Sanger sequencing is used to investigate a short DNA sequence (± 500 bp) after amplification of the region of interest by PCR. After amplification, two different sequence reactions (forward and reverse) are performed and can be used to identify bacterial or fungal species based on the analyses of the sequenced 16S ribosomal DNA (rDNA) and 18S rDNA of the internal transcribed spacer (ITS) region, respectively [2].
One of the disadvantages of Sanger sequencing is that species identification in clinical materials containing more than one species is difficult, if not impossible. Furthermore, the costs and the labour needed for investigating multiple genomic regions of interest makes this method of limited use in modern clinical microbiology laboratories.
Next-generation sequencing (NGS)
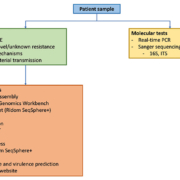
NGS determines the whole genome sequence of different pathogens in one single sequencing run. This technology allows sample multiplexing and, thus, simultaneously provides genomic sequence information on diverse pathogens isolated from different patients. NGS also allows determination of microbial genomes in complex multi-species patient samples by shotgun metagenomics (third generation sequencing) [3]. In comparison to Sanger sequencing, NGS is a considerable improvement owing to the usage of one protocol for all pathogens [4]. A schematic overview of the general workflow used for the sequence analysis in the UMCG is shown in Figure 1.
Using NGS, the whole genome of a pathogen is sequenced in a random way. As benchtop next-generation sequencers can sequence DNA fragments between 100 and 1000 bases, the genome is fragmented before sequencing [5, 6]. Third generation sequencers are an exception to this, as they can handle larger fragments of over 200 kb [2]. NGS requires the preparation of libraries, in which fragments of DNA or RNA are linked to adapters and barcodes. At a later stage, this enables the identification of the sequenced fragments (reads) to the pathogens. After fragmentation, clonal amplification, normalization and a sequencing run is performed. For this, a robust preparation of libraries and standardized protocols are key [3].
Software for data analysis
A huge challenge for the introduction of NGS in a clinical setting is the data analysis. This requires specific software as well as scientific knowledge to interpret the results. There are, so far, only a few user-friendly software packages available to perform data analyses with little bioinformatics knowledge. However, the costs of these software packages is relatively high. However, there a numerous freely available software packages to answer different scientific questions, but knowledge of bioinformatics is often required [2].
After high-throughput sequencing, the reads can be assembled, either by mapping or de novo assembly [2]. Software packages, such as CLC Genomics Workbench (Qiagen), SPAdes and Velvet, can be used for assembly. The genetic relatedness between isolates can be investigated using a gene-by-gene approach using multi-locus sequence typing (MLST), core genome MLST (cgMLST) or whole genome MLST (wgMLST) using SeqSphere+ (Ridom), Bionummerics (Biomérieux), or online tools, such as Enterobase (https://enterobase.warwick.ac.uk) and BIGSdb (http://bigsdb.readthedocs.io). Currently, it is still a matter of debate how many alleles two genomes may differ by to call them genetically related. The same problem applies for comparing two genomes by single nucleotide polymorphism (SNP) typing.
There are a number of web-based tools to perform additional NGS analysis [2]. One of them is the website of the Centre for Genomic Epidemiology (www.genomicepidemiology.org) that can be used for the detection of resistance and virulence genes. Another web-based tool is the Rapid Annotation using Subsystem Technology (RAST) website (http://rast.nmpdr.org) for annotating bacterial genomes.
One of the advantages of web-based tools is that, in general, no knowledge of bioinformatics is necessary. However, a disadvantage may be the lack of tweaking the software settings while performing the analysis. In addition, it may be necessary to confirm the results obtained through web-based tools using other methods [2].
NGS in clinical microbiology
NGS is already applied in several medical microbiology laboratories where it is used for outbreak management, molecular case findings, characterization and surveillance of pathogens, for example [2].
Indeed NGS can be extremely useful in outbreak detection, by monitoring the evolution and dynamics of multi-drug resistant pathogens [7]. A number of studies have highlighted the effectiveness of WGS-based typing for assessing of (newly) emerging pathogens. In our hospital, NGS was used for the characterization of a newly emerging CTX-M-15 producing Klebsiella pneumoniae clone [8]. Transmission of this K. pneumoniae strain between patients has been traced using genomic phylogenetic analysis (Fig. 2). In addition, the study showed the usefulness of a unique marker PCR, in which a clone-specific PCR was developed to investigate the transmission between patients [4].
In addition to tracing and characterizing outbreaks, NGS can be used for the implementation of control measures to avoid the spread of resistance bacteria [9]. An outbreak of a colistin-resistant carbapenemase-producing K. pneumoniae (KPC) with inter-institutional spread in The Netherlands was identified and characterized using NGS and, partially based on these findings, controlled by transferring all positive patients to a separate location [9].
Furthermore, NGS data stored in databases can be used to search retrospectively for molecular case studies. A study from Bathoorn et al. showed that a New Delhi Metallo-?-lactamas-5 (NDM-5)-producing K. pneumoniae was isolated from a Dutch patient. Molecular case findings showed that the Dutch strain is clonally related to strains isolated from four Danish patients in 2014. There was no obvious epidemiological link between the cases in the Dutch and Danish hospitals [10].
These studies and many others highlight the importance of NGS in clinical microbiology. NGS can be used either as a highly discriminatory tool to discriminate between bacterial clones with specific features and to use the information for patient management, infection prevention and evolutionary studies [2] or to characterize bacterial isolates in more detail [8]. Furthermore, web-based databases can be in silico screened retrospectively for the presence of novel (antibiotic-resistance) genes.
Conclusion and outlook
Using NGS, one laboratory protocol can be used to generate sequencing data from samples obtained from different sources. After data analysis, information on the presence of virulence factors and antibiotic resistance genes, as well as other relevant genes are obtained. In addition, NGS makes it possible to standardize typing methods, although cut-off values regarding cgMLST, wgMLST and SNP analysis have to be established internationally in order to distinguish related or unrelated isolates and being able to compare results between laboratories. In the next few years, the role of NGS will surely increase in medical microbiology laboratories, both for research as well as for molecular diagnostic purposes, infection prevention and molecular-epidemiological investigations.
Nonetheless, improvement of the NGS workflow is still needed, focusing on easier and faster ways of library preparation, shorter run-times and further reduction in costs. Furthermore, automatic pipelines for data analyses and easy to use software have to be developed. In addition, the development of proficiency testing panels are important for external quality controls. Only with implementation of the above items at local, (inter)regional and international level will broad use of NGS be allowed in clinical microbiological laboratories for patient and infection control management, including defining a tailor-made antibiotic therapy for each patient, leading to personalized microbiology.
Acknowledgement
A full version of this work is published in the review ‘Application of next generation sequencing in clinical microbiology and infection prevention’, Journal of biotechnology 2017; 243: 16–24.
References
1. Buchan BW, Ledeboer NA. Emerging technologies for the clinical microbiology laboratory. Clin Microbiol Rev 2014; 27(4): 783–822.
2. Deurenberg RH, Bathoorn E, Chlebowicz MA, Couto N, Ferdous M, Garcia-Cobos S, Kooistra-Smid AM, Raangs EC, Rosema S, Veloo AC, Zhou K, Friedrich AW, Rossen JW. Application of next generation sequencing in clinical microbiology and infection prevention. J Biotechnol 2017; 243: 16–24.
3. Head SR, Komori HK, LaMere SA, Whisenant T, Van Nieuwerburgh F, Salomon DR, Ordoukhanian P. Library construction for next-generation sequencing: overviews and challenges. Biotechniques 2014; 56(2): 61–64, 6, 8, passim.
4. Zhou K, Lokate M, Deurenberg RH, Tepper M, Arends JP, Raangs EG, Lo-Ten-Foe J, Grundmann H, Rossen JW, Friedrich AW. Use of whole-genome sequencing to trace, control and characterize the regional expansion of extended-spectrum beta-lactamase producing ST15 Klebsiella pneumoniae. Sci Rep 2016; 6: 20840.
5. Junemann S, Sedlazeck FJ, Prior K, Albersmeier A, John U, Kalinowski J, Mellmann A, Goesmann A, von Haeseler A, Stoye J, Harmsen D. Updating benchtop sequencing performance comparison. Nat Biotechnol 2013; 31(4): 294–296.
6. Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol 2012; 30(5): 434–439.
7. ECDC. Expert opinion on whole genome sequencing for public health surveillance. 2016.
8. Zhou K, Lokate M, Deurenberg RH, Arends J, Lo-Ten Foe J, Grundmann H, Rossen JW, Friedrich AW. Characterization of a CTX-M-15 producing Klebsiella pneumoniae outbreak strain assigned to a novel sequence type (1427). Front Microbiol 2015; 6: 1250.
9. Weterings V, Zhou K, Rossen JW, van Stenis D, Thewessen E, Kluytmans J, Veenemans J. An outbreak of colistin-resistant Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae in the Netherlands (July to December 2013), with inter-institutional spread. Eur J Clin Microbiol Infect Dis 2015; 34(8): 1647–1655.
10. Bathoorn E, Rossen JW, Lokate M, Friedrich AW, Hammerum AM. Isolation of an NDM-5-producing ST16 Klebsiella pneumoniae from a Dutch patient without travel history abroad, August 2015. Euro Surveill 2015; 20(41).
The authors
Sigrid Rosema BSc; Ruud H. Deurenberg PhD; Monica A. Chlebowicz PhD; Silvia García-Cobos PhD; Alida C. M. Veloo PhD; Alexander W. Friedrich MD, PhD; John W. A. Rossen PhD, MMM
Department of Medical Microbiology, University of Groningen, University Medical Center Groningen, The Netherlands
*Corresponding author
E-mail: j.w.a.rossen@rug.nl