Molecular differentiation of ulcerative colitis and Crohn’s colitis: is it achievable?
Differentiating ulcerative colitis from Crohn’s colitis among patients with indeterminate colitis (IC) is a major challenge. The definitive diseases share demographic and clinical features, yet differ in tissue inflammation and damage suggesting distinct mechanisms. Since treatments differ, a molecular diagnostic from accessible clinical samples would greatly benefit IC patients.
by Amanda Williams and Dr Amosy M’Koma
Background
Predominantly, colonic inflammatory bowel disease (IBD), or the colitides, encompasses ulcerative colitis (UC) and Crohn’s colitis (CC) [1, 2], and (when state-of-the-art diagnostic criteria for either are inconclusive) indeterminate colitis (IC) [3]. UC and CC share many demographic and clinical features yet present significant differences in tissue inflammation and damage, suggesting a distinct etiopathogenic trigger [4]. It is believed theoretically that IBD is caused by inappropriate activation of the mucosal immune system against commensal bacteria in the intestinal lumen [4]. Differentiating UC and CC among patients with IC has remained a major challenge in endoscopic precision medicine [5]. Disease unpredictability, treatment side-effects, potential surgery, interim morbidity and acute incapacitation are individual and system burdens [6]. Because treatments for the two diseases are different, identifying phenotype-specific molecular markers would be invaluable for developing diagnostic and prognostic tools, and for precise treatment [7–9].
The need for IC classification into UC and CC is urgent for patients suffering from IBD [10]. Patients diagnosed with IC are young [11], with onset of symptoms before or shortly after the age of 18 years [11, 12] and have an equal gender distribution [13]. This contrasts to UC where there is a male predominance and a mean age of onset at 36–39 years [14]. These figures have persisted despite the introduction of newer diagnostic modalities [15]. Even after long-term follow-up, a substantial number of patients with IC still retain the diagnosis [15]. The continued presence of an IC diagnosis over time supports part of our hypothesis that IBD may represent a spectrum of diseases rather than just two the entities of CC and UC. In order to understand and resolve this challenge, an exclusion tool for differential diagnosis is needed.
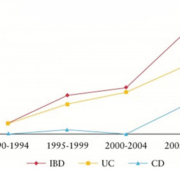
To date there is no diagnostic gold standard tool for IBD. Clinicians use an inexact classification system which combines clinical, endoscopic, radiological, and histopathological techniques in order to diagnose CC and UC [15]. Even with a combination of these methods, IBD patients are mistakenly diagnosed 30% of the time [15], resulting in inappropriate pharmacologic and surgical interventions, with correspondingly significant complications [16]. The most difficult and painstaking post-operative experience is when patients pouch-operated for definitive UC change in their diagnosis to de novo Crohn’s ileitis (CI) of the ileal pouch [15]. Currently, little is known about the molecular differences distinguishing UC and CC [7, 8]. Trends in the IBD field focus on genetic susceptibility, role of normal flora, inflammatory processes, and interactions between normal flora and the immune response [17]. Even though current research is promising [8, 15], there have been no definitive answers to help clinicians differentiate between the two diseases when current diagnostics prove inadequate and result in a diagnosis of IC [3]. Rising incidence and prevalence of IBD (Fig. 1) across the world [18] is accompanied by an increase in cases of IC [11, 19]. It is becoming even more important to find molecular markers of disease to distinguish between CC and UC in patients with IC [7, 8].
Transcriptome analysis
Recently, we have quantitated the global expression profiles of RNA levels using oligonucleotide microarray/genome-wide transcriptome analysis [20, 21] to investigate transcriptional signatures present in colonic tissues obtained from UC and CC mucosa and submucosa. We used genomic data mining from pragmatic studies to demonstrate how biomedical studies can use the technology. By extracting new and useful biomedical knowledge, we hope to develop significant momentum for applications that may have medical diagnostic potential in IBD laboratories. The genomic patterns we noted show greater intensity in CC versus UC, perhaps indicative of a greater degree or different type of inflammation in the tissues underlying the layers [8]. It is possible also that these differing genes may represent candidate biomarkers that could delineate the inflammatory colitides. Specifically, these genes were noted to show greater intensity in the CC submucosa, perhaps indicative of the greater degree or different type of inflammation in the underlying tissue [20, 21]. These studies identified genes involved in inflammatory responses generally overexpressed in IBD and demonstrate that the colonic tissue transcriptomes obtained from UC/CC patients were quite different. The gene sets identified appear to distinguish UC from CC, and may serve as an excellent resource for professionals involved with gene expression data mining in a variety of clinical settings (Table 1).
Proteomics
More recently, we have developed a proteomic approach to delineating UC versus CC. Using histologic mucosal and submucosal tissue layers for analyses, we used MALDI MS for proteomic profiling along with bioinformatics technologies (Fig. 2) [7, 8]. We profiled surgical pathology resections of colonic mucosal and submucosal layers of patients with IBD undergoing colectomy in connection with pouch surgery [restorative proctocolectomy (RPC) and ileal pouch-anal anastomosis (IPAA)] [7, 8, 21]. We identified and compared protein profiles which had the necessary: (1) specificity; (2) sensitivity; (3) discrimination; and (4) predictive capacity to determine the heterogeneity of IBD7, and we were able to delineate UC and CC molecularly [7]. These molecular fingerprints are independent of tissue (mucosa, submucosa, or both) and appear to represent disease-specific markers (Table 1) [7]. Once these markers are further tested, we can potentially develop IBD screening tools which will rely on antibodies to the protein(s) of interest (Fig. 3). The distinction between UC and CC is of the utmost importance when determining candidacy for a pouch surgery [22–24]. Approximately 30% of IBD patients [7] face potential morbidity from an incorrect diagnosis with consequently inappropriate and unnecessary operative surgeries, underscoring the necessity of research efforts aimed at a more accurate diagnosis of the colitides [7, 20].
Peripheral blood biomarkers
In contrast to colon surgical pathology tissue resections, peripheral blood is a much more accessible source of cells that might be used to distinguish between CC and UC. Circulating peripheral blood cytokines are responsible for surveying the body for signs of disease. Cytokines may, therefore, serve as surrogates for disease-induced gene expression as biomarkers of disease status or severity. In pursuit of this, we studied differences in the serum cytokine behaviours between UC and CC patients [9]. We aimed so that, if successful, such analysis could lead to an assay that could be applied as an easy, accurate, affordable, non-invasive and fast screening test. However, although certain cytokines were found to differ between diseases and controls, no cytokine could clearly distinguish UC from CC [9]. An analysis of the literature has shown that although several attempts have been made to define the serum cytokines profile in IBD, the contradictory results of these studies do not indicate the possibility of finding the biomarker(s) among the serum cytokines at this time.
Differential diagnosis and treatment
These studies are highly relevant for creating a molecular differentiator for IC. Curative treatment for UC is often surgical, involving RPC and IPAA [6, 22]. Successful surgery removes the entire diseased colon while preserving bowel evacuation, continence and fertility [22]. This is largely a result of careful patient selection combined with meticulous surgical technique, but most importantly correct diagnosis [16, 22]. Clinical observations and experience suggest that it is difficult to identify patients with CC who are likely to have a successful outcome after RPC and IPAA surgery [6, 16, 23]. Thus, pouch surgery should be widely contraindicated by CC, but be an acceptable intervention for patients with UC and for those with IC who are likely to develop UC.
Despite the increased use of cutting-edge technologies, there is no single, straight- forward explanation for the heterogeneous results, and current approaches still require validation, and subsequently confirmation on patient outcomes in a large-scale clinical cohort.
Conclusion
Our multilevel transcript observations by proteomics and genomics in tissue and blood suggest that the development of a molecular biometric-based tool that can complement the inexact classification system for diagnosis of UC and CC with precision in IBD is still preliminary.
References
1. M’Koma AE, et al. Annual Congress – Digestive Disease Week, Chicago, IL, 2009; M1096 P600
2. Burakoff R. J Clin Gastroenterol. 2004; 38: S41–43.
3. Ballard BR, et al. World J Gastrointest Endos. 2015; 7: 670–674.
4. Podolsky DK. N Engl J Med. 2002; 347: 417–29.
5. North American Society for Pediatric Gastroenterology, Hepatology, and Nutrition, et al. J Pediat Gastroenterol Nutr. 2007; 44: 653–674.
6. Keighley MR. Acta Chir Iugosl. 2000; 47: 27–31.
7. M’Koma AE, et al. Inflamm Bowel Dis. 2011; 17: 875–883.
8. Seeley EH, et al. Proteomics Clin Appl. 2013; 7: 541–549.
9. Korolkova OY, et al. Clin Med Insingts Gastroenterol. 2015: 8: 29–44.
10. Telakis ET. Ann Gastroenterol. 2008; 3: 173–179.
11. Malaty HM, et al. J Pediat Gastroenterol Nutr. 2010; 50: 27–31.
12. Kugathasan S, et al. J Pediatrics 2003; 143: 525–531.
13. Lindberg E, et al. J Pediat Gastroenterol Nutr. 2000; 30: 259–264.
14. Lee KS, et al. Arch Pathol Lab Med. 1979; 103: 173–176.
15. M’Koma AE. World J Gastrointest Surg. 2014; 6: 208–219.
16. Shen B. Inflamm Bowel Dis. 2009; 15: 284–294.
17. Corfield AP, et al. Bioch Soc Trans. 2011; 39: 1057–1060.
18. M’Koma AE. Clin Med Insights Gastroenterol. 2013; 6: 33–47.
19. Malaty HM, et al. Clin Exp Gastroenterol. 2013; 6: 115–121.
20. M’Koma A, et al. Gastroenterology 2010; 138: S-525.
21. M’Koma AE, et al. Oral presentation at the annual congress of The American Society of Colon and Rectal Surgeons, Minneapolis, MN, USA 2010: 117.
22. M’Koma AE, et al. Int J Colorectal Dis. 2007; 22: 1143–1163.
23. Shen B, et al. Inflamm Bowel Dis 2008;14:942–948.
24. Shen B, et al. Clin gastroenterol Hepatol. 2008; 6: 145–158.
The authors
Amanda Williams1 MS; Amosy M’Koma*2,3,4 MD, PhD
1School of Medicine, Meharry Medical College, Nashville, TN, USA
2Department of Biochemistry and Cancer Biology, School of Medicine, Meharry Medical College, Nashville, TN, USA
3Department of Surgery, Vanderbilt University School of Medicine, Nashville, TN, USA
4Vanderbilt-Ingram Cancer Center, Vanderbilt University Medical Center, Nashville, TN, USA
*Corresponding author
E-mail: amkoma@mmc.edu