NGS for inherited skin diseases
Inherited skin diseases can be difficult to assess clinically and often diagnosis relies on multiple laboratory investigations. Traditionally, examination of skin biopsies is followed by biochemical testing and Sanger sequencing of genomic DNA. This approach is labour-intensive, costly and time-consuming. The advent of next-generation sequencing (NGS) methods provides an alternative or complementary approach to making highly accurate diagnoses, but is not without its own challenges.
by J. Lee, Dr A. Salam, Dr T. Takeichi and Prof. J. A. McGrath
Background
The identification of pathogenic mutations in monogenic diseases represents one of the major challenges, and fundamental goals, of early 21st Century human genetics. Most genetic diseases are rare, clinically heterogeneous, and difficult to diagnose – a task made more challenging by disparity in genotype–phenotype correlations, inter- and intra-familial variability, and well as mosaic patterns of disease. It is these hurdles that have led to the advent of Next-Generation DNA Sequencing (NGS); a group of technologies that can improve the speed, accuracy, and cost-efficiency of genetic sequencing, while simultaneously mapping normal variation, and thus furthering our understanding of human genetics in both health and disease. Inherited skin diseases encompass a collection of over 500 clinical entities – with variable structural or inflammatory manifestations that can also affect hair, nails, teeth and certain mucosal surfaces [1]. Individually these disorders are uncommon, but collectively they generate a significant health burden and many diagnostic conundrums.
Traditional approaches to the diagnosis of inherited skin diseases
For patients with inherited skin disorders, the traditional approach to diagnosis is to document a comprehensive patient history, including recording accurate family pedigrees, and noting any consanguinity. The clinician will then go on to perform a physical examination, take clinical photographs, and order laboratory investigations, which often include a skin biopsy. Light microscopy is usually uninformative, and the skin may need to be examined by transmission electron microscopy and immunohistochemistry. Additional blood or urine samples may be need for further diagnostic biochemical studies. Changes in skin structure or protein expression may provide clues to candidate genes, for which polymerase chain reaction primers can be designed and used for Sanger sequencing of genomic DNA. This ‘candidate gene’ approach has proved very useful for several autosomal recessive inherited skin diseases, but is typically unhelpful in most dominant diseases or in those with more subtle changes in skin morphology. Cue the advent of NGS technologies and a different approach to diagnostics, where the challenge in genetic discovery shifts away from the generation of data, to the filtering of relevant data [2, 3].
The impact of NGS
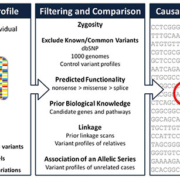
NGS encompasses a number of new technologies that vary in their sequencing protocols, thus determining the type of data produced. The approaches taken vary in template preparation, sequencing and imaging, genome alignment and assembly methods. The methodology is therefore also known as high throughput or massively parallel sequencing due to the ability of NGS to process large volumes of genetic data in a short time, in stark contrast to individual gene screening with Sanger sequencing. Whole-genome sequencing (WGS) and whole-exome sequencing (WES) are the two most commonly used NGS techniques. WGS has the ability to sequence an individual’s entire genome, but at the expense of speed and cost. In contrast, WES uses an array to capture protein-coding regions of the human genome, encompassing ~21,000 genes, which make up less than 2% of the genome. Compared with the 2–3 million variants generated by WGS, the data from WES typically reveals around 25,000 variants. Nevertheless, WES is a more economical option than WGS because ~85% of the pathogenic mutations in monogenic diseases are predicted to be in exons. The plethora of data then has to be filtered, with any potentially disease variant with evidence for causality established (Fig. 1). This process often involves the filtering of variants through databases of previously identified sequences, and cross referencing with known biological or genetic databases, for which considerable bioinformatics support is required: a single WES run can generate one terabyte of data.
Whole-exome sequencing: the possible advantages
The challenge
The key questions for WES in the diagnosis of inherited skin diseases are as follows. (1) Are the new technologies better than what already exist for diagnosing known diseases? (2) Can the new technologies be helpful in resolving unknown diagnoses or discovering new clinical entities? (3) Can the new technologies be introduced into clinical work and overcome any practical obstacles? Emerging data indicate a resounding yes to the first two questions, although the third remains work in progress [4].
Breadth of cover
WES encompasses most of the coding regions of the genome, whereas Sanger sequencing targets a predetermined gene, or part of a gene, between specially designed primers. WES is also efficient for sequencing large genes, such as COL7A1, which encodes type VII collagen. This gene, which is mutated in the blistering disease, dystrophic epidermolysis bullosa, is composed of 118 exons. Conventional Sanger sequencing approaches are based on designing ~72 primer pairs to amplify the COL7A1 exons and flanking introns. Thus the Sanger sequencing approach is therefore laborious and expensive, particularly as COL7A1 contains few recurrent mutations and the gene needs to be screened in its entirety to identify pathogenic mutations.
Genetic diagnosis
WES has emerged as an invaluable tool where a patient’s clinical diagnosis is unclear or erroneous. In this situation, Sanger sequencing of multiple candidate genes is destined to failure and to exhaust both time and resources. WES, on the other hand, can identify known variants in order to make a genetic diagnosis that was not initially considered, as has been demonstrated for subtypes of epidermolysis bullosa, and other inherited skin diseases [5, 6]. Indeed, WES has been used to accurately diagnose inherited skin diseases without any a priori clinical information [7]. The rationale is that more accurate and timely diagnoses offered by WES will allow for earlier targeted therapy and ultimately improved patient care.
Genetic discovery
The value of WES in genetic discovery is evident in the number of inherited skin diseases whose original genetic basis has been informed by WES. Recent examples include the discovery of inherited skin and bowel inflammation resulting from mutations in ADAM17 and EGFR [8, 9]. Given the protean nature of inherited skin diseases, many mutations cannot be anticipated based on clinical phenotype and initial investigations, leaving no candidate gene targets for Sanger sequencing. One pertinent example of the completely unexpected candidate gene is the identification of mutations in EXPH5 [10], which encodes a GTPase effector protein, exophilin-5, in a form of intra-epidermal epidermolysis bullosa – a disease that usually arises as a genetic disorder of keratin. WES is therefore superior to Sanger sequencing in the diagnosis of both novel and genetically heterogeneous conditions.
Cost efficiency
The cost of DNA sequencing has reduced by around 100,000-fold over the last 20 years. Although the technique remains relatively expensive at present (~£900 per sample at King’s College London, 2014 prices), further cost reductions are expected that will soon make WES a more economically viable option than Sanger sequencing, for all but a few disorders in which there are recurrent mutations in a small number of small genes. Even at current costs, however, WES already has advantages over Sanger sequencing for some genes, such as COL7A1, for which the cost of Sanger sequencing is ~£1000 (or greater) in the small number of laboratories that undertake sequencing of this gene.
Considering the patient
The diagnosis of many inherited skin disorders often relies on invasive investigations such as sampling a small piece of skin (punch or ellipse biopsy) (Fig. 2). The procedure involves injection of local anesthetic, which can be painful, and the wound usually heals with a small but evident scar. Occasionally, skin biopsy sites can be complicated by bleeding or infection. WES can be performed using DNA extracted from blood, saliva or tissue samples, and although Sanger sequencing can also be performed on similar templates, for many patients, a skin biopsy would have been necessary to determine the gene(s) for sequencing. Thus WES typically offers a less-invasive approach for the patient.
Variant mapping
Aside from discovering genes and pinpointing mutations in inherited skin diseases, WES also generates a huge amount of other data that can be used to map genetic variation. In the longer term, the dissection of bioinformatics data will lead to a better understanding of the implications of certain variants, refining genotype–phenotype correlation, thus providing insight into individual prognosis, and allowing stratified or personalized medicine and therapeutics.
Whole-exome sequencing: the possible disadvantages
Data analysis
The large quantity of sequencing data generated by WES is potentially also a disadvantage. Before WES can be used in routine clinical practice, fast and efficient filtering techniques must exist to allow clinicians and non-geneticists to interpret WES data and to extract the relevant information in order to manage their patient’s needs. But the plethora of data generated by WES also provides considerably more information beyond the pathogenic mutation itself, including several co-incidental potentially damaging mutations (known as ‘incidental findings’) that are completely unconnected to the primary disease being investigated. What should diagnosticians do with this information? Does it make a difference if the implications are clinically actionable or not? There are clearly several unresolved issues.
Accuracy of data
Given the volume of data produced by WES, it is inevitable that some false positive variants are identified. Most laboratories therefore still elect to confirm mutations via an alternative sequencing platform, generally Sanger sequencing, which is therefore a significant barrier to the routine use of WES in diagnostics. From a technical perspective, NGS methods still need to be improved to cover important regulatory elements such as promoters and enhancers, and poorly annotated parts of the genome. Moreover, if WES is to become a routine diagnostic technique, standardized operating procedures and protocols must be created and implemented. For inherited skin disease diagnostics there would also need to be a realignment of technical wet lab skills (skin microscopy) in favour of computer database and in silico work.
Time to diagnosis
Perhaps the biggest challenge for WES, however, lies in the time it takes to process and analyse a case. For many inherited skin diseases, a rapid diagnosis is often very important to optimize clinical management, for example in neonates with suspected epidermolysis bullosa. The diagnostic approach using skin biopsy assessment followed by Sanger sequencing of candidate genes (implicated by skin biopsy) allows for possible diagnoses to be made within 2 to 3 days. In contrast, the quickest time that WES could be completed (at present) would be a minimum of 5 days, although in practice WES often takes considerably longer to complete and analyse. New platforms to shorten WES protocols are in development, but only when more rapid sample analysis is feasible in a diagnostic lab setting can one really begin to think about wholesale change of diagnostic practice.
Conclusion
Since 2011, WES has proven to be a valuable asset in the diagnosis and discovery of inherited skin diseases. But the adoption of WES into clinical diagnostics diagnosis is still being refined and piloted. WES techniques are constantly being improved to become more accurate, quicker and cost-effective, while enrichment methodologies and sequencing technology become more reproducible and standardized. This progress may allow WES to function independently as the stand alone diagnostic and discovery tool in genetics, negating the need for Sanger sequencing to confirm WES findings. However, as our understanding of the role of non-coding DNA in molecular biology grows, and as WGS is further refined, WES is at risk of being superseded by newer NGS techniques both for genetic discovery diagnostics and prognostics. Innovation looms, but ever it was in molecular genetics.
References
1. Leech SN, Moss C. Br J Dermatol. 2007; 156: 1115–1148.
2. Metzker ML. Genome Res. 2005; 15: 1767–1776.
3. Metzker ML. Nat Rev Genet. 2010; 11: 31–46.
4. Cho RJ, et al. J Invest Dermatol. 2012; 132(E1): E27–28.
5. Takeichi T, et al. Br J Dermatol. 2014; doi: 10.1111/bjd.13190. [Epub ahead of print]
6. Salam A, et al. Matrix Biol. 2013; 33: 35–40.
7. Takeichi T, et al. Exp Dermatol. 2013; 22: 825–831.
8. Blaydon DC, et al. N Engl J Med. 2011; 365: 1502–1508.
9. Campbell P, et al. J Invest Dermatol. 2014; doi: 10.1038/jid.2014.164. [Epub ahead of print].
10. McGrath JA, et al. Am J Hum Genet. 2012; 91: 1115–1121.
The authors
John Lee, Amr Salam BSc, MBChB, MRCP(UK), Takuya Takeichi MD PhD, John A McGrath* MD FRCP
St John’s Institute of Dermatology, King’s College London (Guy’s Campus), London, UK.
*Corresponding author
E-mail: john.mccgrath@kcl.ac.uk