Pharmacogenomics and therapy for hepatitis C virus infection
Pharmacogenomic research has been active in the area of hepatitis C virus infection. Both viral and host polymorphisms are discussed in this review to describe how the genotype information helps predict response to conventional peginterferon alfa and ribavirin therapy as well as more recently recommended direct-acting antivirals, simeprevir and sofosbuvir.
by M. Kawaguchi-Suzuki and Dr R. F. Frye
Hepatitis C virus infection
Hepatitis C virus (HCV) chronically infects 170 million people worldwide [1]. After exposure to HCV, some patients may experience fever, fatigue, dark urine, clay-coloured stool, abdominal pain, loss of appetite, nausea, vomiting, joint pain or jaundice [2]. However, the majority of patients are asymptomatic and do not seek medical attention, leading to the chronic infection rate of 75–85% [2]. HCV infection is a significant burden to society, not only because of the prevalence but also because the infection can lead to severe complications and mortality. Patients infected with HCV can develop chronic liver disease, progressing to advanced fibrosis and cirrhosis and eventually to hepatocellular carcinoma [1]. Additionally, HCV infection is the primary indication for liver transplantation in developed countries [1].
HCV is a blood-borne 9.6 kb positive-sense, single-stranded RNA virus [1, 2]. Typically, the first screening method used to diagnose HCV is the detection of anti-HCV antibodies [3]. Definitive diagnosis of HCV infection is then made by measurements of HCV RNA or ‘viral load’, with a sensitive molecular method having a lower limit of detection <15 IU/mL [3, 4]. Once a decision to treat has been made, the therapeutic goal is to achieve virologic cure or sustained virologic response (SVR) defined as undetectable HCV RNA 12 weeks after the completion of therapy [4]. Recent advancements in HCV treatment now make HCV curable for more patients, which will reduce mortality and liver-related health adverse consequences.
Therapy for HCV infection
Since the identification of HCV in 1989, various treatment regimens have been used to treat HCV infection [1]. Interferon therapy was the first treatment against HCV and then combination therapy with ribavirin (RBV) became available in 1998 [1]. After the introduction of the pegylated formulation of interferon or peginterferon alfa (PEG) in 2001, the dual therapy of PEG and RBV has been the standard care for a decade, until the recent approval of direct-acting antivirals (DAAs) [1]. The first-generation DAAs are the protease inhibitors boceprevir and telaprevir. Subsequently, another protease inhibitor, simeprevir, and a polymerase inhibitor, sofosbuvir, were introduced. The three protease inhibitors were approved in combination with PEG and RBV. However, PEG-free regimens became an option for select patients with the approval of sofosbuvir.
Although the likelihood of achieving SVR has improved with each advance in treatment, HCV infection is a therapeutic area in which large inter-patient variability in response has been observed. In order to predict treatment response and to tailor therapy for individual patients infected with HCV, the extent to which pharmacogenomic biomarkers explain this variability has been examined.
Pharmacogenomics: viral polymorphism
HCV is categorized into genotypes 1–7 and further classified into subtypes a, b, etc., based on sequence divergence [1]. The observed HCV genotype depends largely on the geographic location; genotype 1 accounts for 70% of cases in the Americas, 50–70% of cases in Europe, and 75% of cases in Japan, with genotypes 2 and 3 being the next most prevalent [1]. In contrast, genotypes 3 and 6 have been widely identified in South and Southeast Asia, while genotypes 4 and 5 are most commonly observed in Africa [1]. Genotype 7 was most recently discovered, but its clinical importance has not yet been determined [1]. Identification of the HCV genotype is important for HCV-infected patients because treatment choice, therapy duration, and treatment response depend on the viral genotype. The first-generation DAAs and simeprevir are only indicated for genotype 1 infection. However, sofosbuvir has pan-genotypic activity and is approved for the treatment of genotypes 1, 2, 3, and 4 [5].
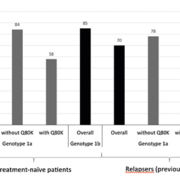
Before the approval of the use of DAAs, HCV therapy targeted the host immune system with the use of PEG. However, since therapy now directly targets the virus itself, various viral polymorphisms that may confer treatment resistance have been reported. The most notable one is the Q80K polymorphism observed in HCV genotype 1a [6]. Findings from the phase 3 QUEST-1, QUEST-2, and PROMISE trials are shown in Figure 1 [6]. QUEST-1 and QUEST-2 were conducted among treatment-naïve patients, whereas PROMISE was a study in patients who relapsed after previous treatment with an interferon-based regimen.
In general, HCV genotype 1a was considered less susceptible to the treatment, but when the data were analysed based on Q80K polymorphism, the SVR rates in genotype 1a were similar to those in genotype 1b if the Q80K polymorphism was absent. However, the SVR rates turned out to be even lower when the polymorphism was present. Based on these data, both prescribing information and clinical guidelines suggest alternative therapy if the Q80K polymorphism is detected in HCV genotype 1a infection [6]. Consequently, Q80K polymorphism testing is recommended by the clinical guidelines in all patients before the initiation of simeprevir, PEG, and RBV triple regimen [4].
Pharmacogenomics: host polymorphism
Earlier, various genome-wide association studies and candidate gene studies found two single nucleotide polymorphisms (SNPs) in the host were associated with SVR in HCV genotype 1 infection after PEG and RBV therapy [7]. The two SNPs, rs12979860 and rs8099917, are located in IFNL3 gene (previously called IL28B) [7]. Rs12979860 CC and rs8099917 TT genotypes have been considered as favourable response genotypes, with SVR rates of around 80% achieved in genotype 1 infection, whereas rs12979860 minor T allele or rs8099917 minor G allele carriers had lower SVR rates of about 20% [8]. SVR rates tend to be higher in HCV genotype 2 and 3 infections with PEG and RBV therapy, compared to those in genotype 1 therapy, and the association of these two SNPs with SVR has not been as strong in genotype 2 and 3 infections [7]. However, similar association of the two IFNL3 SNPs with SVR to that in genotype 1 infection has been shown in genotype 4 infection [7]. Although data has been scarce in other rare genotype infections, the IFNL3 genotype has been one of the strongest predictors of SVR with PEG and RBV therapy [7, 8]. The exact mechanism by which the IFNL3 SNPs affect the phenotype has not been fully elucidated, but baseline differences in the expression level of interferon-stimulated genes have been proposed [9]. Data has been collected with both IFNL3 SNPs, but rs12979860 is more commonly used if a single SNP has to be chosen for research or clinical purpose.
The association of treatment response with the IFNL3 genotype has also been observed with interferon-based DAA therapies. Currently, treatment with boceprevir or telaprevir is not recommended by the guidelines, and most commonly used regiments include simeprevir and/or sofosbuvir [4]. Figure 2 describes SVR rates in treatment-naïve and treatment-experienced patients who were treated with simeprevir or sofosbuvir combined with PEG and RBV as indicated in the prescribing information [5, 6]. Higher SVR rates were consistently observed in patients with the rs12979860 CC genotype, compared to the T allele carriers. However, it should be noted that the difference in SVR rates between the IFNL3 genotypes was comparatively modest with the addition of a DAA to the conventional PEG and RBV therapy. In addition, no significant difference was observed in SVR rates based on the IFNL3 genotype in patients infected with HCV genotype 2 or 3 after an interferon-free regimen of sofosbuvir and RBV [10].
Summary and future directions
Large inter-patient variability exists in response to PEG and RBV therapy, and the IFNL3 genotype has been demonstrated as one of the strongest predictors for SVR, especially in HCV genotype 1 infection. This trend has also been observed in interferon-based DAA therapies. However, if an interferon-free regimen becomes more readily available in the future, with the potential for SVR rates to approach 100%, the IFNL3 genotype may no longer hold clinical utility. However, IFNL3 genotype may still need to be tested during drug development to ensure that investigational agents will have efficacy in patients carrying the variant allele previously associated with an unfavourable treatment response. Additionally, when PEG is eliminated from treatment regimens and only DAAs are combined, cross-resistance may become a concern in the future, especially in patients who failed a DAA regimen previously. Various viral polymorphisms have been detected in protease inhibitors, and cross-resistance can be an issue in this drug class [6, 11, 12]. Viral polymorphisms may play a bigger role and need to be monitored for the future.
References
1. Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med. 2013; 19(7): 837–849.
2. Centers for Disease Control and Prevention. Hepatitis C information for health professionals. 2013; http://www.cdc.gov/hepatitis/hcv/. Accessed January 11, 2014.
3. EASL. EASL Clinical Practice Guidelines: Management of hepatitis C virus infection. J Hepatol. 2014; 60(2): 392–420.
4. AASLD, IDSA, IAS–USA. Recommendations for testing, managing, and treating hepatitis C. 2014; http://www.hcvguidelines.org/.
5. Sovaldi (sofosbuvir) package insert. Gilead Sciences, Inc. 2013.
6. Olysio (simeprevir) package insert. Janssen Therapeutics. 2013.
7. Kawaguchi-Suzuki M, Frye RF. The role of pharmacogenetics in the treatment of chronic hepatitis C infection. Pharmacotherapy. 2014; 34(2): 185–201.
8. Pacanowski M, et al. New genetic discoveries and treatment for hepatitis C. JAMA. 2012; 307(18): 1921–1922.
9. Cariani E, et al. Translating pharmacogenetics into clinical practice: interleukin (IL)28B and inosine triphosphatase (ITPA) polymophisms in hepatitis C virus (HCV) infection. Clin Chem Lab Med. 2011; 49(8): 1247–1256.
10. Jacobson IM, et al. Sofosbuvir for hepatitis C genotype 2 or 3 in patients without treatment options. N Engl J Med. 2013; 368(20): 1867–1877.
11. Victrelis (boceprevir) package insert. Merck & Co., Inc. 2013.
12. Incivek (telaprevir) package insert. Vertex Pharmaceuticals 2013.
The authors
Marina Kawaguchi-Suzuki PharmD, BCPS; Reginald F. Frye* PharmD, PhD, FCCP
Department of Pharmacotherapy and Translational Research,
University of Florida College of Pharmacy, Gainesville,
FL 32610-0486, USA
*Corresponding author
E-mail: frye@cop.ufl.edu