Proteomic approach to investigate ALL biomarkers for early diagnosis and treatment evaluation
The aim of this study was to perform proteomic analysis of serum from pediatric patients with B-cell acute lymphoblastic leukemia (B-ALL) to identify candidate biomarker proteins, for use in early diagnosis and evaluation of treatment. This approach is an alternative to traditional techniques that can investigate the disease from another perspective. Acute lymphoblastic leukemia is the most common malignant cancer in childhood and the symptoms of childhood cancer are difficult to recognize.
by Dr M. de S. Cavalcante, Prof. A. E. Vieira-Neto,
Dr R. de A. Moreira and Dr A. C. de O. Monteiro-Moreira
Background and significance
Acute lymphoblastic leukemia (ALL) is the most common malignant cancer in childhood, and is responsible for approximately 25% of all childhood cancers and 72% of all cases of pediatric leukemia [1]. The current standards for diagnosis of ALL integrate the study of cell morphology, immunophenotyping and genetics/cytogenetics, as described in the classification of lymphoid cancers published by the World Health Organization (WHO) in 2008 [2]. Of lymphoid cancers, as designated using the most recent WHO classification, the purely leukemic presentation, B-lineage ALL (85 %) is the most common [3], and will be addressed in this study. The signs and symptoms of childhood cancer are very challenging to identify, as it is not the first diagnosis to be considered for nonspecific complaints, leading to potential uncertainty in diagnosis. Moreover, children showing the first signs of cancer frequently do not appear severely ill, which may delay diagnosis. In addition, childhood cancer can mimic other common childhood diseases and even normal developmental physiological processes [4]. In the specific case of ALL, early diagnosis and treatment increase the chances of a cure [4].
Future prospects
A label-free proteomic approach was used for the quantitative analysis. Other approaches could also be used in the future, for example it is possible to find studies using RNA interference, mainly silencing expression of specific genes [5]. In our proteomic approach, for each protein, the program ExpressionE selected all corresponding peptides from the samples and compared the intensities of these for relative protein quantification. Using the intensity of a peptide of known quantity, alcohol dehydrogenase (ADH), the program performed self-standardization of data sets. Lists of proteins were then filtered to show only those present in all three repeated injections of each sample, from which an output table was created. This table showed the names, access codes, and expression levels of the proteins, and indicated whether they were upregulated ≥2-fold, downregulated ≤0.5-fold, or whether they did not show significant differences between the groups (unchanged), 0.5 < expression level < 2. The list of proteins generated from three injections of samples in MS, coupled with broad limits used for protein expression levels and serum samples used the controls (non-leukemic pediatric patients) may suggest that the panel of candidate protein biomarkers is clearly increased in the disease state.
Biotechnological resources
Affinity chromatography with α-D-galactose-binding lectin from Artocarpus incisa [6] immobilized on a SepharoseTM 4B gel, combined with identification and quantification of glycoproteins by mass spectrometry, are excellent tools for comparative serum studies. The biomarker pipeline is commonly viewed as a series of preclinical phases: biomarker discovery, and verification before the final clinical evaluation. The comparative analysis results in a list of hundreds of proteins that are differentially expressed between healthy and diseased samples [7]. In this study, the preclinical phase of biomarker discovery was applied and a proteomic analysis of serum samples from pediatric patients with B-ALL was performed, to analyse levels of glycoprotein expression, with the aim of identifying biomarkers to aid in the early diagnosis of B-ALL and to assess the response to induction therapy.
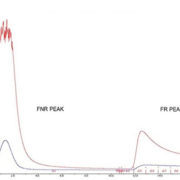
The depletion of high-abundance proteins in serum, human serum albumin (HSA) and IgG, followed by affinity chromatography with the plant lectin Frutalin immobilized on SepharoseTM 4B (Fig. 1), reduced the dynamic range and increased the capacity to identify lower-abundance proteins. The retained fraction (FR) peak containing the protein of interest was concentrated and digested, for later analysis by nano-LC-MS/MS.
Proteomic approach
The study population was composed mainly of children from the lower middle class, who attended a reference hospital for the diagnosis and treatment of childhood cancers in the State of Ceará, Brazil. The study was conducted with the approval of the Research Ethics Committee at the Hospital Infantil Albert Sabin, associated with the Secretary of Health of the State of Ceará. The demographic and clinical data for the patients are summarized in Table 1. The pediatric patients were evaluated at two different times: at diagnosis (B-ALL group; n = 10) and after induction therapy (AIT group; n = 10). Samples of healthy children (Control group; n = 10) were obtained for comparison.
The differentially expressed proteins were used for pathway analysis. Swiss-Prot accession numbers were inserted into the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) software, version 9.05 (available at http://string.embl.de/), with the following analysis parameters: Homo sapiens, confidence level 0.400–0.900, using the active prediction method [8].
Biomarker panel for ALL diagnosis
A panel of protein biomarker candidates has been developed for pre-diagnosis of B-ALL and also provide information that would indicate a favourable response to treatment after induction therapy. In the proteomic analysis, a total of 96 proteins were identified. Leucine-rich alpha-2-glycoprotein 1 (LRG1), Clusterin (CLU), thrombin (F2), heparin cofactor II (SERPIND1), alpha-2-macroglobulin (A2M), alpha-2-antiplasmin (SERPINF2), Alpha-1 antitrypsin (SERPINA1), Complement factor B (CFB) and Complement C3 (C3) were over-expressed in the B-ALL compared to the Control and AIT groups, and were, therefore, identified as candidate biomarkers for early diagnosis of B-ALL. The AIT group showed no significant differences in the expression levels of these proteins compared to the Control group, and did not show any significant change in the level of expression of these proteins, a fact that further reaffirms the presence of these potential biomarkers in a disease state, as all patients achieved complete remission after treatment (Fig. 2). Our results also confirm the important relationship between cancer and phenomena associated with blood coagulation. Several studies have reported that approximately 50% of patients with malignant disease and more than 90% of those that evolve to metastasis present evidence of abnormalities in coagulation and/or fibrinolysis [9–13].
Conclusion
Acute lymphoblastic leukemia is the most common malignant cancer in childhood and this proteomic approach is an alternative to traditional techniques, since the signs and symptoms of childhood cancer are very challenging to identify. LRG1, CLU, F2, SERPIND1, A2M, SERPINF2, SERPINA1, CFB, and C3 were identified as candidate biomarkers for early diagnosis of B-ALL; all were over-expressed in the B-ALL group compared to the Control and AIT groups. The AIT group did not display any significant changes in the expression levels of these proteins, compared to the Control group. All patients in the AIT group achieved complete remission after treatment; this indicates that these biomarkers are only present in the disease state. These candidate biomarkers may improve the pre-diagnosis of B-ALL, which is currently difficult to diagnose in the early stages; the biomarkers may also provide key information on the response to treatment after induction therapy. Further clinical and genomic studies will be important to improve the survival of children with this disease.
Acknowledgements
FINEP, CNPq, RENORBIO-UNIFOR, ALBERT SABIN HOSPITAL
This article is a summary of a paper first published in Biomarker Research: Cavalcante Mde S, Torres-Romero JC, Lobo MD, Moreno FB, Bezerra LP, Lima DS, Matos JC, Moreira Rde A, Monteiro-Moreira AC. A panel of glycoproteins as candidate biomarkers for early diagnosis and treatment evaluation of B-cell acute lymphoblastic leukemia. Biomarker Research 2016; 4: 1 (doi: 10.1186/s40364-016-0055-6) [14].
References
1. Scheurer ME, Bondy ML, Gurney JG. Epidemiology of Childhood Cancer. In: Pizzo PA, Poplack DG, editors. Principles and practice of pediatric oncology, 6th ed, pp2–16. Lippincott Williams and Wilkins 2011.
2. Vardiman JW, Thiele J, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood 2009; 114: 937–951.
3. Chiaretti S, Zini G, Bassan R. Diagnosis and subclassification of acute lymphoblastic leukemia. Mediterr J Hematol Infect Dis. 2014; 6: e2014073.
4. Rodrigues KE, Camargo B. Diagnóstico precoce do câncer infantil: responsabilidade de todos. Rev Assoc Med Bras. 2003; 49: 29–34 (in Portuguese).
5. Trougakos IP, So A, et al. Silencing expression of the clusterin/apolipoprotein j gene in human cancer cells using small interfering RNA induces spontaneous apoptosis, reduced growth ability, and cell sensitization to genotoxic and oxidative stress. Cancer Res. 2004; 64: 1834–1842.
6. Monteiro-Moreira ACO, Pereira HD, et al. Crystallization and preliminary x-ray diffraction studies of Frutalin, an α-D-galactose-binding lectin from Artocarpus incisa seeds. Acta Crystallographica Session F, 2015.
7. Parker CE, Borchers CH. Mass spectrometry based biomarker discovery, verification, and validation–quality assurance and control of protein biomarker assays. Mol Oncol. 2014; 8(4): 840–858.
8. Jensen LJ, Kuhn M, et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009; 37: D412–416.
9. Kwon H-C, Oh SY, et al. Plasma levels of prothrombin fragment F112, D-dimer and prothrombin time correlate with clinical stage and lymph node metastasis in operable gastric cancer patients. Jpn J Clin Oncol. 2008; 38: 2–7.
10. Bick RL. Coagulation abnormalities in malignancy: a review. Semin Thromb Hemost. 1992; 18: 353–372.
11. Luzzatto G, Schafer Al. The prethrombotic state in cancer. Semin Oncol. 1990; 17: 147–159.
12. Nigel O’Connor, Gozzard DI, et al. Haemostatic abnormalities and malignant disease. Lancet 1986; 8: 303–304.
13. Hillen HF. Thrombosis in cancer patients. Ann Oncol. 2000; 11: 273–276.
14. Cavalcante Mde S, Torres-Romero JC, et al. A panel of glycoproteins as candidate biomarkers for early diagnosis and treatment evaluation of B-cell acute lymphoblastic leukemia. Biomarker Research 2016; 4: 1 (doi: 10.1186/s40364-016-0055-6).
The authors
Márcio de Souza Cavalcante1, Antonio Eufrásio Vieira-Neto², Renato de Azevedo Moreira3, Ana Cristina de Oliveira Monteiro-Moreira3*
1Northeast Network of Biotechnology (RENORBIO), State University of Ceará, Ceará, Brazil.
2Center of Experimental Biology (NUBEX), University of Fortaleza (UNIFOR), Ceará, Brazil.
3Department of Biochemistry and Molecular Biology, Federal University of Ceará, Ceará, Brazil.
4Development and Technological Innovation in Drug Program, Federal University of Ceará, Ceará, Brazil
5Reference Center at Children’s Cancer Diagnosis and Adolescents Dr. Murilo Martins, Albert Sabin Hospital, Ceará, Brazil.
*Corresponding author
E-mail: acomoreira@unifor.br