RND efflux pumps in P. aeruginosa: an underestimated resistance mechanism
An adequate initial antibiotic therapy is a key determinant of therapeutic success in Pseudomonas aeruginosa – infected patients. Antibiotic efflux is an underestimated resistance mechanism because it may occur in strains categorized as susceptible. It is rarely or not at all diagnosed in routine laboratories and often masked by high-level resistance mechanisms.
by Dr Laetitia Avrain, Dr Pascal Mertens and Professor Françoise Van Bambeke
P. aeruginosa: state of the art of antibiotic susceptibility
P. aeruginosa is a Gram-negative bacterium recognized as a major cause of infections in hospitalized patients or in patients with impaired defences as observed in burn wounds or cystic fibrosis. In spite of improved hygiene measures, the risk of infection by P. aeruginosa in ICU remains high (infection incidence > 30/100 patients hospitalized in ICU). P. aeruginosa infections are associated with mortality rates as high as 30 % to 50 % in bacteremia [1] and up to 70% in patients with nosocomial pneumonia [2].
Yet, empirical selection of antibiotics is made difficult by the continuously evolving resistance of P. aeruginosa to antibiotics, notably due to the emergence of Multi Drug Resistance (MDR) phenotype (R ≥ 3 antibiotic classes). The MDR status of the strain as well as an initial inappropriate treatment negatively influence patient outcome [3].
Acquired high level resistance is due to the acquisition of genes coding for aminoglycoside-modifying enzymes or beta-lactamases, or to mutations in fluoroquinolone targets. Intrinsic antibiotic resistance is due to low outer membrane permeability mediated either by under production of the oprD porin, or by the expression of multidrug resistance efflux pumps. The genome of P. aeruginosa codes for numerous efflux pumps, among which MexAB-OprM and MexXY-oprM are of first clinical importance due to their large prevalence in clinical strains and their ability to expel several classes of chemically-unrelated antibiotics.
RND efflux pumps in P. aeruginosa
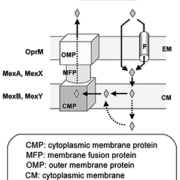
The main efflux pumps in P. aeruginosa belong to the Resistance-Nodulation-Division (RND) superfamily, which uses proton motive force as energy source. They constitute a tri-partite system, composed of an integral cytoplasmic membrane drug-proton transporter, an outer membrane channel and a periplasmic fusion protein linking the two other proteins. This assembly allows expelling the substrate from the inner membrane directly to the extracellular medium [Fig. 1, reproduced from [4]].
Ten efflux systems have been characterized in P. aeruginosa, among which MexAB-OprM and MeXY-OprM are constitutively expressed at a basal level in wild-type strains (expression of MeXY-OprM being however much lower than that of MexAB-OprM). Both systems are inducible when exposed to antibiotic substrates. The other systems (MexCD-OprJ, MexEF-OprN, MexJK, MexGHI-OpmD, MexVW, MexPQ-OpmE, MexMN, and TriABC are not expressed in wild type strains but may contribute to antibiotic or biocide resistance when expressed in resistant strains [5].
Antipseudomonal antibiotics released by P. aeruginosa multidrug efflux systems
RND efflux systems release multiple antimicrobials components including first-line antipseudomonal antibiotics such as β-lactams and β-lactamase inhibitors, fluoroquinolones, aminoglycosides [Table 1]. More specifically MexAB-OprM transports β-lactams fluoroquinolones, macrolides, tetracyclines, trimethoprim, sulfamides and chloramphenicol; MexXY-OprM, aminoglycosides, fluoroquinolones, macrolides, and tetracyclines; MexCD-oprJ, some β-lactams, fluoroquinolones, macrolides, tetracyclines, trimethoprim and chloramphenicol, and MexEF-OprN, fluoroquinolones, trimethoprim and chloramphenicol. The latter is also involved in resistance to meropenem and doripenem, but this may rather result from the fact that the OrpD porin is downregulated in strains expressing this efflux system.
Colistin, the last resort drug for MDR P. aeruginosa, is not substrate for these efflux pumps. Thus, efflux is responsible for multidrug resistance, a single pump being able to transport several classes of drugs while at the same time some redundancy exists among transporters, fluoroquinolones for example being universal substrates for the main efflux systems. Moreover, the subsequent reduction in antibiotic concentration inside the bacteria may help selecting high level resistance mechanisms, in particular target mutations [6].
Over-expression of efflux pumps: impact on antimicrobial susceptibility
A study published in 2010 examined the impact of antibiotic treatment on the susceptibility of P. aeruginosa, by collecting successive isolates from ICU patients at the time of diagnosis of infection and during treatment [7]. Globally, mean minimum inhibitory concentration (MIC) values increased after exposure to antibiotics, with statistically significant effects being observed for amikacin, ciprofloxacin, cefepime, meropenem and piperacillin/tazobactam, bringing mean MICs to values higher than the EUCAST susceptibility breakpoints. Three quarters of the isolates showed a moderate elevation of the MIC (≤16X initial MIC), suggesting the involvement of low to moderate levels resistance mechanisms as those affecting membrane permeability [Fig 2, reproduced from [7]].
The analysis of the expression of efflux pumps in this collection revealed that a high proportion of the strains (34 %) did overexpress MexAB-OprM and MexXY-OprM in the initial isolate, but that this proportion further increased during the antibiotic treatment, with about two third of the strain overexpressing at least one of these efflux systems [Fig.3, adapted from [8]].
Diagnosis of efflux in clinical laboratory
Because efflux in P. aeruginosa almost always co-operates with other mechanisms of resistance, differential diagnosis by phenotypic antimicrobial analysis is complex, high levels resistance mechanisms masking the effect of the expression of efflux systems on MICs. Moreover, efflux pumps can be overexpressed during treatment, which may explain therapeutic failures with antibiotics that are considered active based on the original determination of the susceptibility profile.
Resistance by efflux can be detected using Efflux Pumps Inhibitors (EPI), which revert MICs to those strains that do not express efflux systems. Among them MC-207,110 (phenylalanine arginyl beta-naphthylamide) is a broad spectrum inhibitor that has been widely used in vitro to investigate the impact of efflux on susceptibility to antibiotics of P. aeruginosa. Inhibitors specific of a given transporter are also under investigation. Yet, in MDR strains with additional resistance mechanisms, EPI do not allow restoring antibiotic activity, which may lead to false-negative results [9].
In this context, molecular methods appear as the only way to evidence the expression of efflux pumps in clinical isolates. Immunoblotting methods were developed first but were rapidly replaced by Reverse Transcriptase quantitative PCR (RT-qPCR) due to its higher specificity and rapidity. RT-qPCRs were thus developed to detect and quantify the expression of the genes coding for the different proteins of a given RND pump. This method remains applicable whatever the other resistance mechanisms present in the clinical strain and can thus be applied in clinical laboratories. Typically, a 2-fold increase in the expression of mexA and mexB genes causes a decrease in antibiotic susceptibility, while overexpression of mexX needs to be higher (≥ 5-fold) to increase MIC values. This low level of overexpression implies that all the steps for RT-qPCR should be carefully standardized [10]. The commercial mex Q-TesT kit includes two housekeeping genes to standardize the RT-qPCR and facilitates the interpretation of mexA and mexX genes expression of clinical Pseudomonas aeruginosa strains in comparison to wild type strain PAO1.
Conclusion
Resistance by efflux has now well been characterized in specialized laboratories but is still rarely or not at all diagnosed in routine laboratories. The complexity of resistance in P. aeruginosa with MDR phenotypes and the lack of diagnostic tools are probably the main reasons why this mechanism is neglected. Because this resistance mechanism can also contribute to therapeutic failures, accurate diagnosis is of prime importance for selecting adequate therapy.
References
1. Aliaga L, Mediavilla JD, et al. A clinical index predicting mortality with Pseudomonas aeruginosa bacteraemia. J Med Microbiol 2002; 51(7): 615-619.
2. Alp E, Guven M, et al. Incidence, risk factors and mortality of nosocomial pneumonia in intensive care units: a prospective study. Ann Clin Microbiol Antimicrob 2004; 3: 17.
3. Hirsch EB, Tam VH. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev Pharmacoecon Outcomes Res 2010; 10(4): 441-451.
4. Mesaros N, Van Bambeke F, et al. L’efflux actif des antibiotiques et la résistance bactérienne: état de la question et implications. La lettre de l’infectiologue 2005; (4): 117-126.
5. Lister PD, Wolter DJ, et al. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 2009; 22(4): 582-610.
6. Zhanel GG, Hoban DJ, et al. Role of efflux mechanisms on fluoroquinolone resistance in Streptococcus pneumoniae and Pseudomonas aeruginosa. Int J Antimicrob Agents 2004; 24(6): 529-535.
7. Riou M, Carbonnelle S, et al. In vivo development of antimicrobial resistance in Pseudomonas aeruginosa strains isolated from the lower respiratory tract of Intensive Care Unit patients with nosocomial pneumonia and receiving antipseudomonal therapy. Int J Antimicrob Agents 2010; 36(6): 513-522.
8. Riou M, Avrain L, et al. Influence of antibiotic treatments on gene expression of RND efflux pumps in successive isolates of Pseudomonas aeruginosa collected from patients with nosocomial pneumonia hospitalized in Intensive Care Units from Belgian Teaching Hospitals. ECCMID, 10-13 April 2010, Vienna, Austria. P780.
9. Van Bambeke F, Pages JM, et al. Inhibitors of bacterial efflux pumps as adjuvants in antibiotic treatments and diagnostic tools for detection of resistance by efflux. Recent Pat Antiinfect Drug Discov 2006; 1(2): 157-175.
10. Avrain L, Hocquet D, et al. Pre-Real-Time PCR steps standardization for appropriate interpretation of mexA and mexX gene expression by mex Q-Test in P. aeruginosa. ECCMID, 10-13 April 2010, Vienna, Austria. P590.
The authors
Laetitia Avrain PhD1*, Pascal Mertens PhD1 and Françoise Van Bambeke, Professor, Maître de Recherche FNRS, PhD2
1 Coris BioConcept, Gembloux, Belgium
2 Molecular and cellular pharmacology,
Louvain Drug Research Institute, Université catholique de Louvain, Brussels, Belgium
*Corresponding author
E-mail: laetitia.avrain@corisbio.com