The role of expressed prostatic secretion biomarkers in prostate cancer screening/surveillance
Expressed prostatic secretion (EPS) is obtained after prostatic message by milking the urethra and collecting fluid directly from the urethral meatus. Clinicians have previously used this biospecimen primarily for the diagnosis of urinary infections. More recently it has been recognized as a rich source of biomarkers of prostate malignancy. Many of those biomarkers are accessible only in EPS, where it has shown a unique potential in both prostate cancer screening and active surveillance programmes.
by Dr J. Linehan, Dr J. Yamzon, Prof. T. Wilson and Prof. S. Smith
Current risk assessment tools
An estimated 238,590 new cases of prostate cancer will be diagnosed in the United States during 2013. A substantial proportion of these patients are considered ‘low-risk’ for progression and may be best served with conservative management. Yet many will undergo definitive treatment by means of radiotherapy or radical prostatectomy, both of which have risks of life-altering side effects. This huge – and ultimately unsustainable – pattern of over-treatment has incited a shift in practice patterns toward initial conservative management known as Active Surveillance (AS), the premise of which is to minimize unnecessary treatment by performing close monitoring. Should there be signs of tumour progression upon recurring reassessment, then treatment can be offered with curative intent. Currently, our ability to discern one’s risk for harbouring an indolent form of prostate cancer utilizes clinical parameters that lack predictive ability. Serum PSA level, clinical stage determined on digital rectal examination (DRE), and tumour grade determined upon biopsy, known as the Gleason Score (GS), are currently used to stratify the risk of tumour progression, although with only limited accuracy. This is evidenced by studies of men eligible for AS who are found to have more advanced disease upon treatment with radical prostatectomy [1] despite meeting AS criteria using the standard criteria. Tools based on the biology of prostate cancer offer a potential platform for improved risk stratification, and hence allow more appropriate levels of intervention.
Invasive and non-invasive testing
Current efforts in biomarker discovery and validation stand to complement the existing tools that gauge eligibility for AS. Several tools, such as Oncotype DX-Prostate® (Genomic Health) and Prolaris® (Myriad Genetics), are commercially available and generate risk scores for harbouring more advanced disease. These rely on extraction of RNA from tumour tissue, most commonly taken from the biopsy specimen that may not contain the sufficient amounts of cancer tissue needed to perform these proprietary tests. AS protocols require periodic repeat biopsies of the tumour, which may have potential complications of bleeding, infection, and pain. Thus, a less invasive method of assessment is ideal. Tools reliant on serum and urine specimens are attractive in that they are more readily obtained. Serum tests, such as free-to-total PSA ratios, the 4K Score (OPKO Health), and the Prostate Health Index (Beckman-Coulter), show promise in refining prostate cancer diagnosis, but these rely on proteins shed from the tumour into circulation. Urine based tests like PCA3 (Gen-Probe) or DNA based tests like ConfirmMDx (MDxHealth) rely on proteins shed into the urine after prostate massage, or DNA extracted from biopsy specimens. These latter tests are utilized in the clinical scenario where prostate cancer remains undiagnosed despite high suspicion by standard parameters.
The biological function of the prostate gland is the production of seminal fluid critical to reproduction. It contains DNA, RNA, proteins and metabolites that have the potential to serve as biomarkers useful in prostate cancer diagnosis and risk assessment. Its contents can be retrieved by two non-invasive methods. The first, variously called a post-DRE urine or post-massage urine, is obtained as a urine sample collected immediately after an attentive DRE. The second, termed prostatic fluid or expressed prostatic secretion (EPS) is obtained after prostatic massage by milking the urethra and collecting fluid directly from the urethral meatus. Neither specimen requires invasive techniques and both contain significant numbers of prostate cancer cells and prostate cancer biomarkers [2–5]. Each has advantages and disadvantages. Post-DRE urine is relatively straightforward to collect; however, the urine volume, and cell lysis that occurs in urine, can significantly dilute the informative cellular material and hinder the recovery of the informative components of the fluid. EPS specimens require more effort to collect but are undiluted and do not promote cell lysis or block activity measurements. Most clinicians would prefer to collect post-DRE urine samples, but the extra effort required to collect EPS could be justified because a number of useful biomarkers cannot be assessed in post-DRE urine. Patient acceptance of both biopsy and EPS collection are improved dramatically by the administration of valium prior to biopsy. Moreover, EPS collection prior to biopsy can be performed in a single clinical visit.
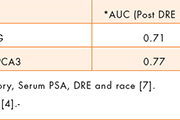
Both EPS and post-DRE urine specimens have proven successful in prostate cancer detection using gene expression biomarkers, where PCA3 and TMPRSS2:ERG fusions have proven effective. Here, sufficient quantities of RNA are present in either specimen, permitting roughly equivalent improvements in performance in the prediction of biopsy outcome (Table 1). The moderate improvement in test performance seen in EPS compared to post-DRE urine appears to be attributable to the need for normalization to PSA messenger RNA measured in the same specimen [6]. This requirement is unique to post-DRE urine as a specimen since urine volume collection is not constant, making it necessary to gauge the informative RNA signals against a fixed RNA signal. The choice of PSA messenger RNA in this application tends to suppress the informative signals since more of it is released into the urine from regions of prostate cancer where the basement membrane is compromised. Moreover it is not possible to measure the volume of the prostatic fluid after it is diluted by the variable urine catch.
DNA methylation is much more difficult to quantify, since DNA is present in much lower concentration in cells, and the required chemical treatments of the isolated DNA results in a severe reduction of the DNA available for quantitative PCR amplification. It has only been reported to be effective in the prediction of biopsy outcome in EPS [4], where the combination of hypermethylation at RARß, RASSF1 and GSTP1performed as well as PCA3 as a single marker.
The field defect and prostate function
Prostate functionality becomes an important diagnostic tool when one considers the extensive field defect that characterizes the disease [8]. Field defects are best defined as functional aberrations that extend beyond the morphologically recognizable tumour in otherwise histologically normal tissue. The concept dates to the early 1950s and has been shown to involve altered DNA methylation patterns extending several millimeters from the prostate tumour. In prostate tumorigenesis it has recently been documented that extensive chromosome rearrangements occur in single steps involving chromothripsis and chromoplexy [9, 10]. In this view the field defect represents clonal progeny of an earlier stage in tumour evolution from which additional chromothripsis and chromoplexy events generate the malignancy. Since most reports of hypermethyled loci fall at or extremely near fragile sites [11], DNA methylation alterations may be linked to chromoplexy and chromothripsis, making it possible to detect cancer by looking for early alterations DNA methylation patterning. Mitochondrial DNA aberrations also appear to track prostate tumour evolution and the field defect includes mitochondrial DNA deletions [12].
Each of these findings suggests that capacity of the prostate gland to produce functional prostatic fluid will be regionally compromised. Genome wide aberrations in DNA methylation coupled with the aberrant expression of transcription factors, such as ERG, could indicate wholesale aberrations in gene expression, while the mitochondrial deletions may underlie alterations in the unique metabolic pathways present in the normal gland. The field defect around larger or more highly evolved tumours could in fact affect the entire gland and can be expected to significantly alter not only the nature but also the amount of prostatic fluid that the gland can produce.
Anecdotally, surgeons collecting EPS by the non-invasive procedure prior to prostatectomy or by squeezing the excised gland just after prostatectomy note that a prostate with a larger tumour burden yields less fluid than a gland bearing small tumour foci, suggesting that the measured volume of the recoverable fluid should be an effective biomarker for prostate cancer. This was demonstrated in recent studies with patients who are eligible for active surveillance by various criteria but were treated with robot assisted radical prostatectomy instead.
For these studies, we used a biomarker set that measures the secretion capacity of the prostate gland. This set includes recovered EPS volume in microlitres and total recovered RNA in nanograms. It is measured along with pre-operative serum PSA value as baseline in multivariate logistic regression analysis (Fig. 1).
Future directions
Given the utility of EPS biomarkers in predicting the presence of occult extracapsular extension in an AS cohort, we believe that the advantages of EPS in assessing the functionality of the gland make it uniquely suited for initial risk stratification in determining eligibility for AS and also in monitoring the disease during surveillance. One of the most perplexing issues in this area is detecting misclassification. In preliminary work in this area (Wittig et al. in prepartion) we noted that more than 40% of patients initially scored as GS 6 in an NCCN cohort can be occult GS 7 comprising largely GS 3+4 patterns with relatively fewer pattern 4+3 GS 7s. Although GS 7 patients are referred for definitive treatment this data suggests that many have not progressed during AS. None of the available biomarkers that we have tested in EPS thus far (TMPRSS2:ERG, PCA3, Secretion Capacity, TXNRD1 or PSA-mRNA) reliably detect this relatively subtle change in the GS. Misclassification recognized at repeat biopsy is generally accepted as an error of initial biopsy undersampling. One can question the clinical significance of such a subtle nuance and its affect on the grander outcomes of disease metastasis and prostate cancer death. Biopsy misclassification is yet another example of the under-performance of standard parameters (like the GS) and the need for biomarker based determinations of disease aggressiveness.
Several new biomarkers are evaluable exclusively in EPS. One example is PSA activity. PSA is a proteolytic enzyme that cannot be assayed in urine specimens, although the assay may function in urine sediment. It is active in EPS and has been shown to be inversely proportional to tumour aggressiveness [13] . Another biomarker with potential in EPS is citrate, which has been shown to be an indicator of prostate cancer in seminal fluid. This marker [14] reflects the overall health of the gland, since the unique metabolism of the normal prostate is designed to secrete citrate into the prostatic fluid. A third biomarker approachable only in EPS is zinc [15]. This ion is taken up preferentially in normal prostate where it blocks the degradation of citrate by inhibiting aconitase in the citric acid cycle thus contributing to the normal function of the gland in citrate secretion.
In summary, the suboptimal performance of the current clinicopatholigic parameters utilized in prostate cancer diagnosis and risk stratification drive the need for additional tools. Biomarkers examined in EPS offer the additional dimension of prostate-function assessment through a non-invasive platform that has great potential to improve prostate cancer diagnosis, risk stratification, and AS.
Abbreviations
AS, active surveillance
AUC, area under curve
DRE, digital rectal examination
EPS, expressed prostatic secretion
GS, Gleason Score
PSA, prostate-specific antigen
ROC, receiver operating characteristic
References
1. Conti SL, Dall’era M, Fradet V, Cowan JE, Simko J, Carroll PR. Pathological outcomes of candidates for active surveillance of prostate cancer. J Urol. 2009; 181: 1628–1633 (Discussion pp. 1624–1633).
2. Groskopf J, Aubin SM, Deras IL, Blase A, Bodrug S, Clark C. APTIMA PCA3 molecular urine test: development of a method to aid in the diagnosis of prostate cancer. Clin Chem. 2006; 52: 1089–1095.
3. Crocitto LE, Korns D, Kretzner L, Shevchuk T, Blair SL, Wilson TG, Ramin, SA, Kawachi MH, Smith SS. Prostate cancer molecular markers GSTP1 and hTERT in expressed prostatic secretions as predictors of biopsy results. Urology 2004; 64: 821–825.
4. Clark JP, Munson KW, Gu JW, Lamparska-Kupsik K, Chan KG, Yoshida JS, Kawachi MH, Crocitto LE, Wilson TG, et al. Performance of a single assay for both type III and type VI TMPRSS2:ERG fusions in non-invasive prediction of prostate biopsy outcome. Clin Chem. 2008; 54: 2007–2017.
5. Laxman B, Tomlins SA, Mehra R, Morris DS, Wang L, Helgeson BE, Shah RB, Rubin MA, Wei JT, Chinnaiyan AM. Noninvasive detection of TMPRSS2:ERG fusion transcripts in the urine of men with prostate cancer. Neoplasia 2006; 8: 885–888.
6. Whelan C, Crocitto L, Kawachi M, Chan K, Smith D, Wilson T, Smith S. The influence of PSA-RNA yield on the analysis of expressed prostatic secretions (EPS) for prostate cancer diagnosis. Can J Urol. 2013; 20: 6597–6602.
7. Tomlins SA, Aubin SM, Siddiqui J, Lonigro RJ, Sefton-Miller L, Miick S, Williamsen S, Hodge P, Meinke J, et al. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci Transl Med. 2011; 3: 94ra72.
8. Mehrotra J, Varde S, Wang H, Chiu H, Vargo J, Gray K, Nagle RB, Neri JR, Mazumder A. Quantitative, spatial resolution of the epigenetic field effect in prostate cancer. Prostate 2008; 68: 152–160.
9. Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature 2012; 482: 53–58.
10. Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, Park K, Kitabayashi N, MacDonald TY, et al. Punctuated evolution of prostate cancer genomes. Cell 2013; 153: 666–677.
11. Smith SS. Maintaining the unmethylated state. Clin Epigenetics 2013; 5: 17.
12. Maki J, Robinson K, Reguly B, Alexander J, Wittock R, Aguirre A, Diamandis EP, Escott N, Skehan A, et al. Mitochondrial genome deletion aids in the identification of false- and true-negative prostate needle core biopsy specimens. Am J Clin Pathol. 2008; 129: 57–66.
13. Ahrens MJ, Bertin PA, Vonesh EF, Meade TJ, Catalona WJ, Georganopoulou D. PSA enzymatic activity: A new biomarker for assessing prostate cancer aggressiveness. Prostate 2013; 73: 1731–1737.
14. Kline EE, Treat EG, Averna TA, Davis MS, Smith AY, Sillerud LO. Citrate concentrations in human seminal fluid and expressed prostatic fluid determined via 1H nuclear magnetic resonance spectroscopy outperform prostate specific antigen in prostate cancer detection. J Urol. 2006; 176: 2274–2279.
15. Zaichick V, Sviridova TV, Zaichick SV. Zinc in the human prostate gland: normal, hyperplastic and cancerous. Int Urol Nephrol. 1997; 29: 565–574.
The authors
Jennifer Linehan MD, Jonathan Yamzon MD, Timothy Wilson MD, and Steven Smith* PhD
NanoLab, Division of Urology, City of Hope, Duarte, California, USA
*Corresponding author
E-mail: ssmith@coh.org