Warfarin: a case for pharmacogenomics testing
The ability to use genetic information to inform clinical decision making has emerged as a new tool in clinical practice, with noteworthy examples across many area of medicine. Cardiology and anticoagulation in particular have led the way in the translation of genetic findings into actionable clinical recommendations, spurred by the addition of genetically guided dosing in the drug label for warfarin by the FDA. This review covers the pharmacogenomics related to warfarin therapy.
by Dr Minoli Perera
Pharmacogenomics is primarily aimed at identifying genetic variation that influences inter-individual differences in drug response. The guiding principle is “the right drug, at the right dose for the right person”. Its application promises to enable targeted drug administration, improve therapeutic outcomes, and inform drug development. Pharmacogenomic insights have also improved our understanding of the underlying pathways and mechanisms behind adverse drug reactions. Such adverse reactions account for approximately 100 000 deaths per year in the US, and markedly increase healthcare costs. Advances made over the last 30 years in molecular biology, molecular medicine, and genomics have had a major impact on the development of pharmacogenomics.
Currently a variety of approaches are used to associate genetic variants associated with drug response. Commonly used strategies include candidate gene approach and genome-wide association studies (GWAS). Candidate gene studies investigate single nucleotide polymorphisms (SNPs) that are correlated to drug response. These studies are usually restricted to genes or SNPs that have been shown to be involved in the pathway of drug action or drug clearance. Genome-wide association studies investigate up to 5 million SNPs spaced throughout the genome which are genotyped to identify genetic variants with the drug phenotype in an unbiased fashion. Each of these methods has advantages and disadvantages. Genome-wide studies comprehensively cover the entire genome, but their power to detect moderate associations is greatly limited by the multiple testing burden, which is a requirement for correction for false-positive associations. The candidate gene approach narrows the focus to a few important SNPs and therefore has higher power, but may miss the real causative SNP, and require a priori knowledge for the selection of SNPs/genes to study.
Both these methods have yielded important clinical findings that can immediately be used to reduce the incidence of adverse effects (many of which have been added to drug labels). Notable examples such as the SLCO1B1*5 polymorphism (associated with myopathy with statin use) and CYP2C19*2 (associated with clopidogrel non-response) have shown clinically meaningful outcomes related to genetic variants. However, the translation of these findings has been slower in coming and has many clinicians wondering about the utility of this new technology.
The “test case” for pharmacogenetics was thought to be pharmacogenetically guided warfarin dosing. In this review we will cover the genetic polymorphism effecting warfarin dose requirements and the currently available diagnostic tests, as a case study for the implementation of pharmacogenetics in the clinic.
Genes that affect warfarin dose
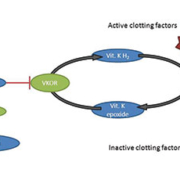
Numerous studies, predominately conducted in Caucasian and Asian populations, demonstrate that the CYP2C9 and VKORC1 genotypes contribute significantly to warfarin dose variability [1–3]. The role of these gene products can be seen in Figure 1. The CYP2C9 enzyme metabolises the more active S-enantiomer of warfarin to inactive 7-hydroxywarfarin. Warfarin inhibits the enzyme VKOR (encoded by the gene VKORC1) to prevent conversion of Vitamin K epoxide to its reduced form necessary for activation of the clotting factors II, VII, IX and X. Thus, SNPs in the CYP2C9 gene affect warfarin pharmacokinetics, whereas variation in the VKORC1 gene impacts warfarin pharmacodynamics.
The most extensively studied CYP2C9 variants are the CYP2C9*2 and CYP2C9*3 alleles, which lead to significant reductions in CYP2C9 activity. Compared with the people without this genotype, carriers of the *2 or *3 genotypes have S-warfarin clearance reduced to between 40 to 90% or normal levels. As a result, significantly lower doses are usually needed in individuals with a CYP2C9*2 or CYP2C9*3 allele. The CYP2C9 genotype is also implicated in the risk of bleeding during warfarin therapy, especially during the warfarin initiation period [4].
The VKORC1 genotype was originally recognised for causing warfarin resistance through mutations in the gene-coding region. More recently, common VKORC1 SNPs occurring in gene-regulatory regions and underlying usual warfarin dose variability were discovered [3]. Two such SNPs, one in the promoter region (-1639G>A) and one in intron 1 (1173C>T), show the strongest association and possible functional effects [5]. Thus, the majority of warfarin pharmacogenetic studies have focused on one of these two SNPs, which are in strong linkage disequilibrium across populations (meaning they are inherited together and strongly associated with each other). This means that only one of these SNPs needs to be taken into account for pharmacogenetic dosing of warfarin. Most investigators chose the VKORC1 -1639G>A as the predictive SNP in warfarin pharmacogenetic studies. This SNP explains approximately 20–28% of the overall variability in dose requirements in Caucasians, but only 5–7% of the variability in African–Americans, mainly due to the difference in allele frequency between populations [6, 7]. Unlike CYP2C9, the VKORC1 genotype does not appear to affect the risk of bleeding with warfarin treatment[4].
These findings have been confirmed in several genome-wide association studies (GWAS) in Caucasians and Asian individuals, showing that VKORC1 -1639G>A, CYP2C9*2 and CYP2C9*3 polymorphisms are the primary genetic determinants of warfarin dose requirements in these populations. The combination of VKORC1 -1639G>A, CYP2C9 (*2 and *3) and clinical factors (e.g., age, sex, weight and amiodarone use) explains approximately 55% of the total variance in warfarin maintenance dose in Caucasians, but only about 25% among African–Americans. With the exception of the CYP4F2 genotype, found in a GWAS study of Swedish patients [8], no other genetic variant has met genome-wide significance for association with warfarin dose requirements. Both genetic and non-genetic variables have been included in dosing algorithms that can be used to predict dose, such as WarfarinDosing.org.
Warfarin genetic testing and guidelines
Insurance companies consider genetic testing for genetic variants in CYP2C19 related to clopidogrel response and the HLA-B* 1502 allele for prediction of adverse effects related to carbamazepine as “medically necessary” and may therefore cover the cost of these tests. The same does not hold for warfarin testing, which is considered investigational and will only be covered in the context of a clinical trial.
In 2007, the US Food and Drug Administration modified the package insert for warfarin to include information on the relationship of safe and effective dosage to SNPs in CYP2C9 and VKORC1, including a table of recommended doses for each genotype combination [Table 1]. These recommendations give the range of doses that should be considered when dosing a patient that is a carrier of any of the tested SNPs. However, there is increasing evidence that additional alleles outside of CYP2C9*2 and *3 and VKORC1 -1639G/A may play a role in warfarin dose response. These SNPs are not included in the FDA dose recommendations and not all tests cover all these additional variants.
Currently, four warfarin pharmacogenetic tests are available as in vitro diagnostic devices [shown in Table 2]. All of these tests genotype for three loci: CYP2C9*2, CYP2C9*3 and one VKORC1 -1639G/A or 1173C/T (both of which give equivalent information because of the afore mentioned LD in all populations), with some including other known genetic variants that are associated with warfarin dose. All of the tests can be completed in 8 hours, including DNA extraction, with the fastest ones providing genotype results in less than 2 hours.
The Clinical Pharmacogenetics Implementation Consortium (CPIC) recently published guidelines on how to interpret and apply genetic test results to adjust warfarin doses [9]. These guidelines do not address when to order a genetic test, but rather how to dose warfarin when genetic test results are available. The guidelines strongly support the use of genetic information to guide warfarin dosing when genotype is known and recommend using either the International Warfarin Pharmacogenetics Consortium (IWPC) or Gage algorithm to do so.
Although the availability of FDA-cleared devices for warfarin pharmacogenetic testing makes genotype-guided warfarin initiation possible, several barriers to clinical adoption remain. First, many medical centres do not have warfarin pharmacogenetic testing available. In a recent survey, only 20% of hospitals in North America have testing available on site, suggesting the majority of the hospitals rely on outside commercial clinical laboratories. This outsourcing may make genotype-guided warfarin initiation impractical because of 3–7 days of turnaround time. Second, no professional organisation endorses warfarin pharmacogenetic testing in its guidelines because of the lack of the clinical utility data. Inclusion of a testing recommendation in professional guidelines has been identified as a factor influencing reimbursement of new technology. As such, the Centers for Medicare and Medicaid Services (CMS) and many commercial insurance plans generally do not reimburse the cost of testing ($300–500). Because of these barriers, warfarin pharmacogenetic testing is performed mainly for research purposes and for patients willing to pay the cost.
Future perspective and conclusions
There are substantial and convincing data supporting the clinical and analytic validity of warfarin pharmacogenetics. The CYP2C9 and VKORC1 genes are the primary determinants of warfarin dose requirements. There are several FDA-cleared tests available for CYP2C9 and VKORC1 genotyping. However, genotype-guided warfarin dosing has not yet become a reality in most medical centres despite the wealth of data supporting genetic influences of warfarin dose requirements. Many clinicians and third party payers are awaiting evidence of clinical utility and cost-effectiveness before adopting genetic testing for anticoagulation management in the clinic setting. Results from ongoing clinical trials (such as the NIH-sponsored COAG trial) are expected to address these issues and will likely determine the course of genotype-guided anticoagulant therapy. Whether pharmacogenetics will have a role in the treatment with newer anticoagulant agents has yet to be determined. However, the pharmacogenetics with these anticoagulants could be of great importance given the unavailability of routine monitoring parameters with these agents.
References
1. Wadelius M, et al. Blood 2009; 113: 784–792.
2. Klein TE, et al. N Engl J Med 2009; 360: 753–764.
3. Rieder MJ, et al. N Engl J Med 2005; 352: 2285–2293.
4. Limdi NA, et al. Clinical pharmacology and therapeutics 2008; 83: 312–321.
5. Wang D, et al. Blood 2008; 112: 1013–1021.
6. Cavallari LH, et al. Clin Pharmacol Ther 2010; 87: 459–464.
7. Perera MA, et al. Clin Pharmacol Ther 2011; 89: 408–415.
8. Takeuchi F, et al. PLoS Genet 2009; 5: e1000433.
9. Johnson JA, et al. Clinical pharmacology and therapeutics 2011; 90: 625–629.
10. Coumadin package insert. 2007. (Accessed October, 2007, at http://www.bms.com/cgi-bin/anybin.pl?sql=PI_SEQ=91.)
The author
Minoli A Perera, PharmD., PhD
Knapp Center for Biomedical Discovery
Room 3220B, University of Chicago, 900 E.
57th Street, Chicago, IL 60637, USA
E-mail: mperera@bsd.uchicago.edu