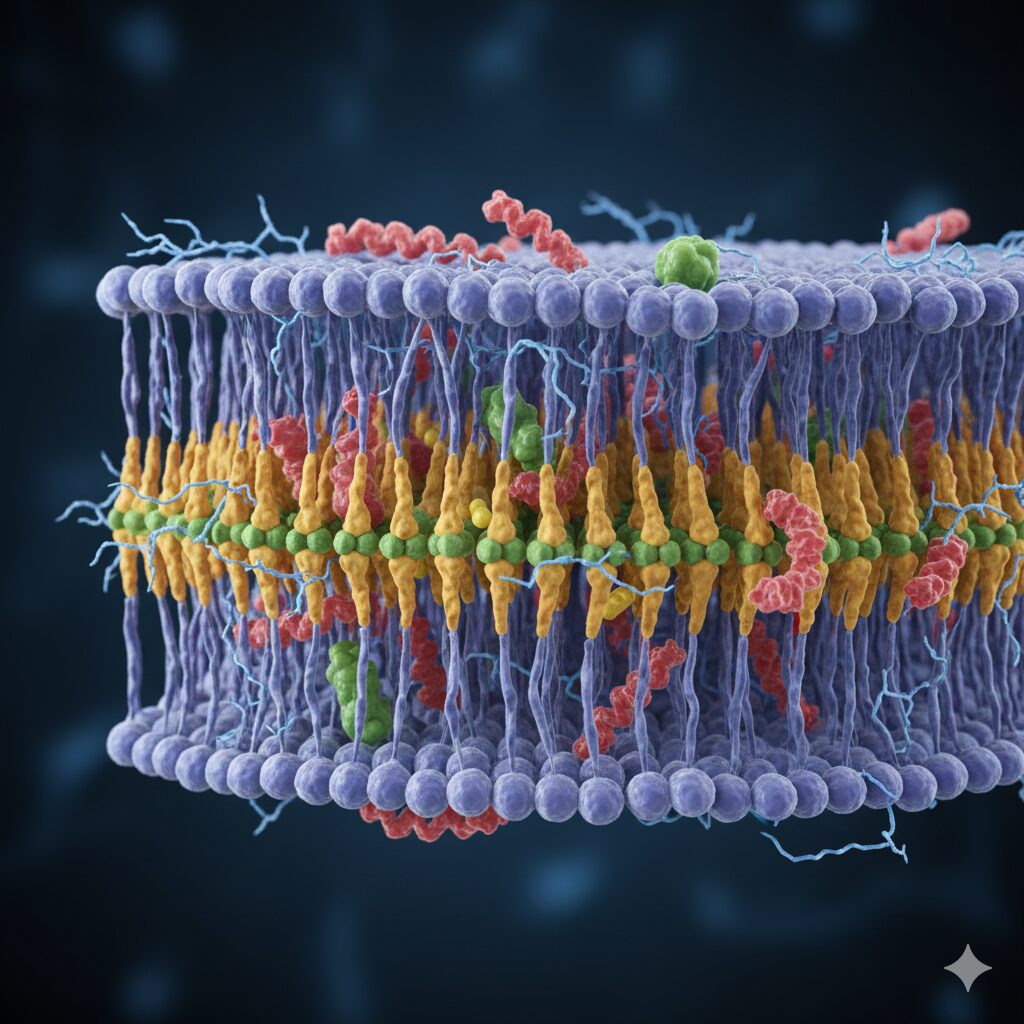
Computational design reveals structural principles governing membrane protein stability
Scientists at Scripps Research have developed a computational approach for designing synthetic membrane proteins, uncovering critical atomiclevel interactions that govern protein folding and stability within cellular lipid environments. The methodology offers new insights into disease-related protein dysfunction and potential therapeutic targets. Membrane proteins constitute essential molecular machinery for cellular function, mediating substance transport, signal transmission, catalytic activity, and intercellular adhesion. Dysfunction in these proteins underlies numerous pathologies, including cancer, establishing them as priority pharmaceutical targets. However, their integration within lipid bilayers has historically complicated structural and functional analysis.
Designer proteins illuminate structural motifs
The research team, led by Assistant Professor Marco Mravic from the Department of Integrative Structural and Computational Biology, focused on a conserved structural motif characterised by small amino acids recurring at seven-residue intervals throughout membrane-spanning protein chains. This pattern positions these amino acids identically on alternate helical turns, suggesting functional significance in helix-helix interactions and membrane organisation.
“Billions and billions of dollars a year are going into making molecules that target membrane proteins to alter their behaviour and combat disease, but in order to modulate these proteins, it helps to first understand how they work,” says Mravic. “Our study uncovered some new rules of sequence and atomic arrangements inside membrane proteins that are essential for them to function.”
The investigators hypothesised that these motifs represent interaction sites facilitating helix binding and architectural organisation within membrane folds. To test this, they employed computational design to generate idealised synthetic versions amenable to laboratory characterisation, circumventing the inherent instability of natural membrane proteins extracted from cellular contexts.
Software-driven sequence optimisation
First author Kiana Golden developed software to identify amino acid sequences containing the target motif, subsequently designing optimised synthetic membrane proteins with enhanced stability. Experimental production of these proteins confirmed predicted folding patterns, validating the hypothesis that the motifs create stabilising interactions between adjacent helices within the lipid environment.
“We found that the motif’s stability was driven by an unusual type of hydrogen bond that’s typically very weak, but when the motif is repeated, these weak hydrogen bonds all add up to make a very stable interaction,” explains Golden, who conducted the work as an undergraduate at UCSD and is now a graduate student at Princeton University. “This type of hydrogen bond is rare in the natural world, so it was really surprising that this is largely what’s driving this motif to occur, and that biology has evolved to use it within specific motifs andpan structures across nature.”
Remarkably, proteins engineered with optimal motif sequences demonstrated exceptional thermal stability, maintaining structural integrity even under boiling conditions.
Therapeutic and diagnostic implications
The structural principles revealed through this work provide a framework for identifying pathogenic mutations affecting membrane protein stability. The demonstrated capability of the computational platform to generate highly stable protein complexes within lipid environments opens avenues for designing therapeutic molecules that directly target membrane proteins in their native cellular context.
“Our approach vastly accelerates what we can discover about the inner workings of membrane proteins and how to make better therapies,” notes Mravic.
The methodology represents a significant advancement in membrane protein research, offering a systematic approach to dissecting the atomic-level determinants of protein behaviour within lipid bilayers. By enabling the design of stable, tractable model systems, the technique circumvents traditional experimental limitations and facilitates mechanistic investigation of membrane protein folding, stability, and function.
The findings, published in Proceedings of the National Academy of Sciences on 7 October 2025, establish design principles for a widespread structural motif and demonstrate the potential of computational protein design in advancing understanding of membrane protein biology.
The research received support from the National Institutes of Health, the Diekman Family Graduate Fellowship, the ARCS Fellowship, and the UCSD McNair Scholars Programme.
Reference:
Golden, K., Avarvarei, C., Anderson, C. T., et. al. (2025). Design principles of the common Gly-X6-Gly membrane protein building block. Proceedings of the National Academy of Sciences, 122. https://doi.org/10.1073/pnas.2503134122