Korean researchers develop ultra-precise CRISPR technique that dramatically improves detection of rare cancer mutations
Researchers have engineered a highly accurate CRISPR variant that removes normal DNA before sequencing, enabling detection of circulating tumour DNA at levels as low as 0.005%. The MUTE-Seq method showed promising results in early-stage lung and pancreatic cancer patients, where conventional liquid biopsy often fails.
A new CRISPR-based liquid biopsy technique has demonstrated remarkable sensitivity in detecting extremely rare cancer mutations, potentially transforming early cancer diagnosis and treatment monitoring. The method, developed by researchers at Korea University College of Medicine and collaborators, uses an engineered enzyme to selectively eliminate normal DNA before genetic sequencing, thereby dramatically amplifying the signal from circulating tumour DNA (ctDNA).
The research, published in Advanced Materials, addresses a fundamental limitation of current liquid biopsy approaches: the exceptionally low concentrations of tumour-derived DNA fragments circulating in blood, particularly during early-stage disease. In some early cancers, ctDNA can constitute as little as 0.01% of total cell-free DNA, rendering detection extraordinarily challenging with conventional next-generation sequencing (NGS).
Engineering precision at the molecular level
At the heart of the advance lies FnCas9-AF2, a rationally engineered variant of Francisella novicida Cas9. The researchers systematically modified specific amino acids within the enzyme’s structure to enhance its ability to discriminate between perfectly matched DNA sequences and those containing even single-base mismatches. “We engineered a highly precise advanced-fidelity FnCas9 variant, named FnCas9-AF2, to effectively discriminate single-base mismatches at all positions of the single guide RNA target sequences,” the authors explain in their paper. This precision represents a substantial improvement over existing high-fidelity CRISPR variants such as SpCas9-HF1 and eSpCas9.
Genome-wide analysis using Digenome-seq revealed that FnCas9-AF2 exhibited near-zero off-target cleavage activity, even when tested simultaneously at multiple genomic locations. Whilst conventional SpCas9 produced hundreds of off-target cuts, FnCas9-AF2 demonstrated exceptional specificity with undetectable off-target effects in several test scenarios.
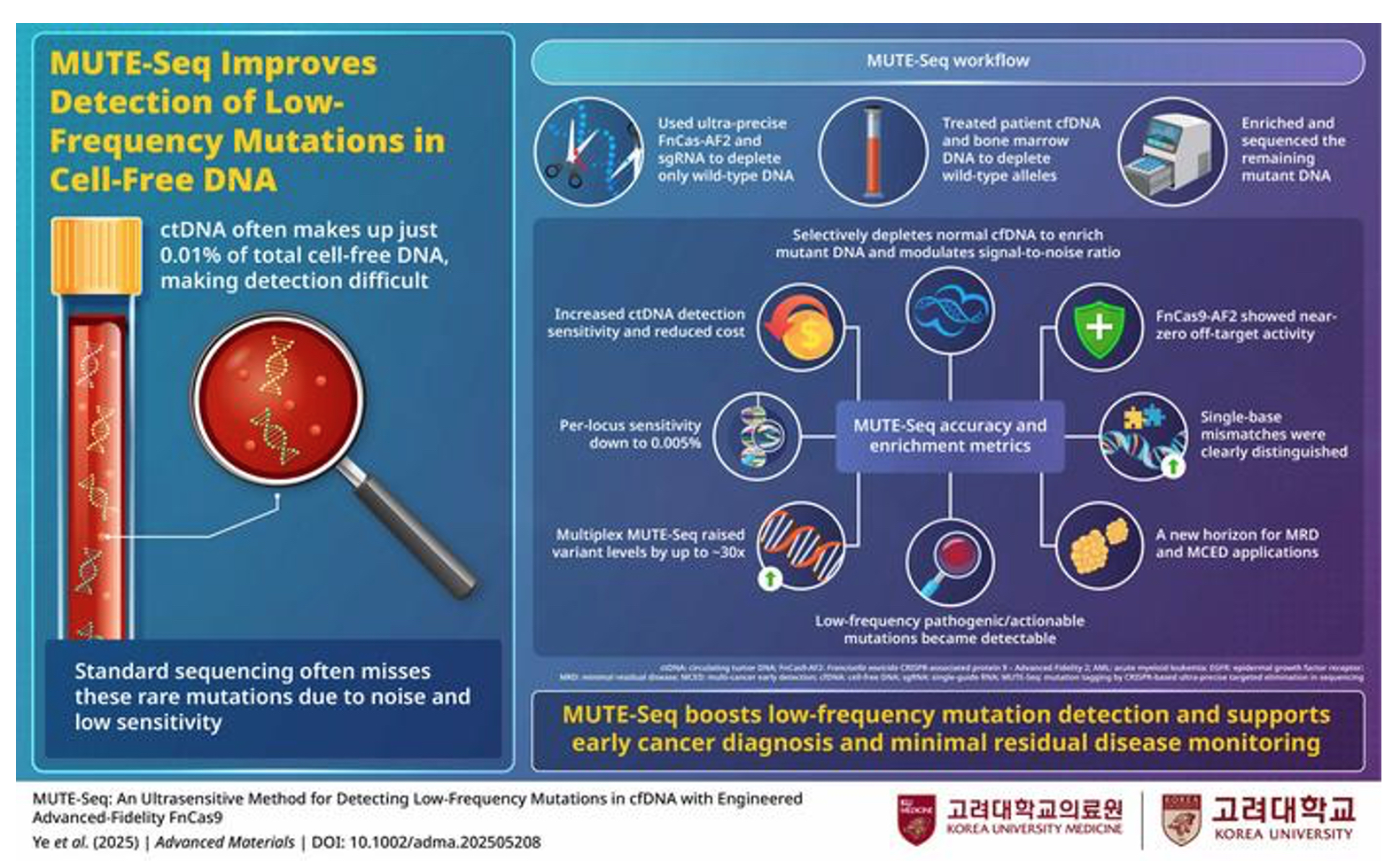
MUTE-Seq uses ultra-precise FnCas9-AF2 to remove wild-type DNA, sharply boosting detection of rare cancer mutations down to 0.005% VAF
(© Professor Junseok W Hur, Korea University College of Medicine, Korea)
Clinical validation across multiple cancer types
The team applied their method – termed MUTE-Seq (Mutation tagging by CRISPR-based Ultra-precise Targeted Elimination in Sequencing) – to clinical samples from patients with acute myeloid leukaemia (AML), non-small cell lung cancer (NSCLC), and pancreatic cancer.
In eight AML patients undergoing minimal residual disease (MRD) monitoring, MUTE-Seq substantially increased the detected variant allele frequencies (VAFs) of NRAS mutations. Samples that initially showed mutations at 3-17% VAF by conventional methods yielded significantly higher detection rates after wild-type depletion, enabling clearer identification of residual disease.
For NSCLC patients, the researchers designed a multiplexed approach targeting 323 clinically relevant mutation sites. When comparing tissue biopsy results with plasma samples from ten patients across disease stages, MUTE-Seq achieved 90.9% sensitivity – substantially higher than the 54.5% sensitivity of conventional sequencing. Notably, the method achieved 100% concordance in stage I patients, where ctDNA levels are typically lowest.
In 20 pancreatic cancer patients who had undergone surgery, most at early stages, MUTE-Seq detected 24 of 29 tissue-confirmed mutations in cell-free DNA samples, achieving 82.8% sensitivity with 100% specificity.
Limits of detection and practical implications
Performance testing with reference materials revealed that MUTE-Seq consistently increased VAFs by 20- to 60-fold compared with controls. Using 50 ng of input DNA – a clinically realistic amount given typical plasma yields – the method achieved a 95% limit of detection of 0.034% VAF, closely matching theoretical predictions.
Professor Junseok W Hur, corresponding author and director of the Centre for Genomic R&D at Korea University Anam Hospital, emphasised the clinical potential: “Our findings suggest that the MUTE-Seq method has considerable potential for developing diagnosis panels aimed at detecting multiple low-frequency ctDNA for MCED [multi-cancer early detection], CDx [companion diagnostics], or MRD monitoring.”
The technique offers several practical advantages. By depleting wild-type DNA before sequencing, it effectively reduces background noise without requiring ultra-deep sequencing or complex unique molecular identifier (UMI) systems, potentially reducing costs whilst improving sensitivity. The method can be integrated into existing laboratory workflows and combined with standard bioinformatic pipelines.
The authors conclude: “We anticipate that the FnCas9-AF2-based MUTE-Seq could offer a valuable clinical tool to facilitate improved molecular diagnosis, prognosis evaluation, and treatment planning for cancers in various stages.”
Reference:
Ye, S., Kim, J.-S., Kim, M., et. al. (2025). MUTE-Seq: An ultrasensitive method for detecting low-frequency mutations in cfDNA with engineered advanced-fidelity FnCas9. Advanced Materials, 37(47), e2505208. https://doi.org/10.1002/adma.202505208