LC-MS/MS for profiling of urinary eicosanoids
The eicosanoids are a family of lipid mediators of pain and inflammation involved in multiple pathologies. Urine has become a useful, readily accessible biofluid for monitoring the endogenous synthesis of these metabolites. The clinical interest of eicosanoids requires their targeted determination, which, in turn, warrants the development of high-throughput analytical methodology such as liquid chromatography–tandem mass spectrometry methods.
By Dr Cristina Gómez
Introduction
Eicosanoids are bioactive lipid mediators produced by the enzymatic and/or non-enzymatic oxidation of arachidonic acid (5,8,11,14-eicosatetraenoic acid), an abundant cell membrane component. Three different oxidative pathways, comprising of cyclo-oxygenase, lipoxygenase and cytochromes P450, enzymatically oxidize arachidonic acid which is converted to different eicosanoids based on the pathway involved (Fig. 1) [1–4]. These signalling mediators typically are not stored within cells but rather synthesized de novo from membrane-released arachidonic acid as required, when cells are activated by mechanical trauma or by specific cytokine, growth factor, and other stimuli. They are found in biological fluids and tissues at low concentrations. They play a fundamental role in promoting and modulating inflammation, providing both pro-inflammatory signals and terminating the inflammatory process, as well as maintaining tissue and vascular homeostasis. Disruption of the homeostasis of eicosanoids is closely related to a range of inflammatory pathophysiological conditions (fever, nephritis, cardiovascular diseases, inflammation, allergy, HIV and cancer) and act as a regulator of important processes (e.g. smooth muscle tone, platelet aggregation and vascular permeability) (1–4).
The levels of lipid mediators in activated cells may rapidly increase by several orders of magnitude compared to baseline. These signalling mediators act locally, but are rapidly removed by the circulation, meta-bolized and transported to the systemic circulation as a mixture of primary and secondary metabolites (2). The circulating metabolites are efficiently cleared by the kidney, and excreted into the urine. Many eicosanoid metabolites are frequently used as biomarkers owing to their involvement in diseases and pathophysiological conditions.
Biomarkers could be defined as compounds found in biological fluids or tissues indicating the presence of an abnormal or significantly changed condition. A biomarker can, for example, be used for disease detection, response to therapy, or to monitor exposure to toxicological compounds or conditions. The compound can either be present in higher or lower amounts than what is considered a normal range. The measurement of eicosanoid metabolites in the urine reflect activation of the different biosynthetic pathways, e.g. analysis of 8-iso-prostaglandin (PG)F2α as a biomarker for oxidative stress, or PGE2 as a biomarker involved in inflammation. Accordingly, the analysis and quantification of these compounds in biological samples has emerged as the most practical and reliable method to assess in vivo production of primary eicosanoids.
Quantification of eicosanoids in urine
To enhance our understanding of disease progression and eicosanoid concentration, measurement of the complete profile of produced eicosanoids is essential. Given the clinical interest in eicosanoids and the complexity of their responses to biological stimuli, it is necessary to systematically monitor the changes in their concentrations in various tissues and biological fluids. During the early inflammatory phase, eicosanoids are excreted by inflammatory cells and can increase up to 100-fold in local concentration [2]. This requires sensitive, selective and reproducible methods for their quantification.
Measurements of eicosanoids in the blood are difficult owing to low circulating levels (both primary eicosanoids and their metabolites are present in blood and urine at concentrations in the picomolar to nanomolar range), rapid hepatic and renal clearance, and induction of biosynthesis during sampling. Urine is, therefore, an optimal non-invasive biofluid for monitoring eicosanoid level, readily accessible biofluid for monitoring the endogenous synthesis of these metabolites and may be interpreted by consideration of the physiological context. The primary eicosanoids present in urine may be derived from the kidneys, whereas the urinary eicosanoid metabolites most likely reflect systemic eicosanoid metabolites [1, 2]. The quantification of primary eicosanoids and their metabolites in urine is sufficient for the assessment of both renal as well as systemic production of eicosanoids. Their quantification can serve as systematic indicators of pathological processes in the vascular, respiratory, and other cellular systems [5–7]. Reliable measurement of selected eicosanoids levels could benefit the selection of precision biomarkers assisting disease prognosis, progression and pharma-codynamic assessment.
The thorough evaluation of the eicosanoid metabolome is useful for understanding the physiology and pathology behind these processes. The utility of urinary profiling to detect inflammatory responses associated with, for example, allergen provocation has been demonstrated in several studies. As an example of the application, changes in urinary metabolites can be quantified and they reflect the local reactions in the airways after an inhalation challenge [6].
LC-MS/MS method for quantification of eicosanoids
Mass spectrometry (MS) metabolite pro-filing approaches provide a useful combination of omics-scale screening, while still focusing on biological pathways that are relevant to the pathology of interest. Historically, gas chromatography (GC)-MS and GC-tandem MS (MS/MS) have played a central role both in the identification and in the quantification of eicosanoids in biological samples. Those methods often require a complex and time-consuming sample extraction and derivatization. Due to great advances in liquid chromatography (LC)-MS/MS technology, especially over the past two decades, this relatively new approach is increasingly used both in biomedical and life sciences, and in clinical chemistry. Up to now, most reported methods examine these pathways in isolation; few studies have examined in depth the urinary eicosanoid profile for large-scale profiling of most of the major pathways at the population level [5, 7].
The quantification of eicosanoids and their metabolites in biological samples confronts an analytical challenge, even though a number of methodologies/techniques have been developed. The major difficulties encountered are related to the oxidation of eicosanoids and their low quantities in biological matrices, which requires sensitive, selective and reproducible methods for their quantification. Besides, eicosanoids are structurally similar, many of them are isomeric compounds that share the same parent mass and also the same fragmentation pattern, such as PGE2, PGD2 and 13, 14‐dihydro‐15‐keto‐PGE2. Therefore, these compounds have to be chromatographically resolved, i.e. it is necessary to achieve sufficient chromatographic sepa-ration to discriminate individual eicosanoids with a high degree of specificity.
Several targeted methods for qualitative and/or quantitative determination of selected panels of eicosanoids in different biofluids have been published. A recent paper reported the development of an eicosanoid profiling method which was designed with the intent to achieve the necessary reproducibility and precision for large-scale molecular phenotyping studies and to enable the direct comparison of urinary excretion levels between independent clinical studies [6]. The method for the extraction and quantification of 32 eicosanoid urinary metabolites by LC-MS/MS covers the major synthetic pathways including prostaglandins, leukotrienes and isoprostanes (Fig. 1). In the LC optimization, column temperature and gradients were adjusted to accomplish complete separation of the critical separation pairs. After LC parameters were optimized, MS parameters were optimized to attain the lowest possible detection limit. Multiple reaction monitoring transitions were selected to yield the greatest selectivity and sensitivity. To improve the quantitative accuracy, deuterated internal standards were selected to compensate for differences in the extraction and ionization efficiency due to differences in chemical structure and chromatographic elution. Matrix effects, suppression or enhancement of the ionization efficiency, were also evaluated. In addition, the method was optimized in terms of low urine volume consumption, broad metabolic pathway coverage, and long-term precision (6).
The resulting method was highly reproducible, repeatable and stable across multiple years of analysis, and is, therefore, well suited for applications in molecular phenotyping studies, drug trials, other clinical investigations, and epidemiological monitoring of responses to exposures. The method presented some advantages with respect to previously published methods, with a focus on formatting the method to be suitable for large-scale analyses and clinical trials [6]. Previous methods were limited in metabolic coverage, required laborious, solvent intensive liquid–liquid extraction, long run-times, time-consuming absorbance measurements, large extraction volume, and/or enzymatic hydrolysis. The workflow defined monitors the majority of the eicosanoid pathways (PGF2α, PGE2, PGI2, PGD2 pathways, isoprostanes, thromboxane and leukotrienes pathways), that have established clinical or physiological functions. Further-more, one of the main advantages is the minimum required volume for analysis with the complete workflow, which simplifies planning of clinical studies and improves on previous methods in terms of simplicity, metabolic coverage and precision. Future application of this method might contribute to a better understanding of the role and relevance of eicosanoid metabolites for inflammatory diseases and conditions.
Summary
LC-MS/MS methods are being used to identify, monitor and quantify eicosanoids, signalling molecules involved in inflammatory processes and is informative for mechanistic insight into numerous pathologies. The described panel covers the major eicosanoid urinary metabolites including the prostaglandins, thromboxanes, leukotrienes, and isoprostanes. The novel method was validated in human urine and was demonstrated to be a simple, robust and sensitive method for quantification of urinary eicosanoids from different pathways by LC-MS/MS applicable in clinical studies.
References
1. Dennis EA, Norris PC. Eicosanoid storm in infection and inflammation. Nat Rev Immunol 2015; 15(8): 511–523.
2. Funk CD. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science 2001; 294(5548): 1871–1875.
3. Milne GL, Yin H, Hardy KD, Davies SS, Roberts LJ. Isoprostane generation and function. Chem Rev 2011; 111(10): 5973–5996.
4. Shimizu T. Lipid mediators in health and disease: enzymes and receptors as therapeutic targets for the regulation of immunity and inflammation. Annu Rev Pharmacol Toxicol 2009; 49(1): 123–150.
5. Sterz K, Scherer G, Ecker J. A simple and robust UPLC-SRM/MS method to quantify urinary eicosanoids. J Lipid Res 2012; 53(5): 1026–1036.
6. Gómez C, Gonzalez-Riano C, Barbas C, Kolmert J, Hyung Ryu M, Carlsten C, Dahlén SE, Wheelock CE. Quantitative metabolic profiling of urinary eicosanoids for clinical phenotyping. J Lipid Res 2019; 60(6): 1164–1173.
7. Sasaki A, Fukuda H, Shiida N, Tanaka N, Furugen A, Ogura J, Shuto S, Mano N, Yamaguchi H. Determination of ω-6 and ω-3 PUFA metabolites in human urine samples using UPLC/MS/MS. Anal Bioanal Chem 2015; 407(6): 1625–1639.
The author
Cristina Gómez PhD
Division of Molecular and Systems Toxicology, Department of Pharmaceutical Sciences,
University of Basel, Switzerland
E-mail: Cristina.gomezcastella@unibas.ch
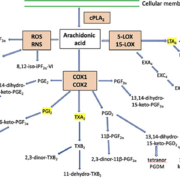
Figure 1 (a)(b). Schematic of the eicosanoid metabolic cascade displaying the urinary metabolites included in the current platform.
Arachidonic acid can be metabolized via lipoxygenase, cyclooxygenase, and oxidative stress (ROS, RNS) pathways. The chromatogram and gradient elution pattern demonstrate the separation of the eicosanoid metabolites.
cPLA2, cytosolic phospholipases A2; COX, cyclooxygenase; EX, eoxin (14,15-disubstituted analogues of the 5,6-disubstituted leukotrienes); LOX, lipoxygenase; LT, leukotriene; PG, prostaglandin; RNS, reactive nitrogen species; ROS reactive oxygen species; TX, thromboxane.