Automated image analysis personalizes colorectal cancer prognosis
The current prognostic staging of colorectal cancer (CRC) by the tumour, node, metastasis method, alongside the minimal core data set, provides good prognostic information for patient populations but is less accurate for the individual. Reporting of additional histopathological features can improve individualized prognostic staging, but manual microscopic surveillance often results in observer variability and there is a lack of consensus on standardized quantification methods. Automated image analysis can standardize the quantification of prognostic features, can personalize CRC prognosis and augment clinical staging.
by Dr Peter Caie
Introduction
Colorectal cancer (CRC) incidence is extremely high in the developed world, where it is the third most common cancer in men and women in both the UK and USA. There will be an estimated 134 500 new CRC cases diagnosed, and just under 50 000 deaths will have been caused by the disease, in the USA alone. Although the incidence rate has decreased only slightly in the last decade, the change in mortality has dropped significantly between 2000 and 2010, with ~50% of patients across all disease stages surviving for at least 5 years post-diagnosis. Reasons for the decrease in mortality include life style change (47% of CRC cases in the UK could be prevented from healthier lifestyle choice) early detection of disease (e.g. home testing such as fecal occult blood kits), targeted therapy as the result of ‘omics’ research, novel prognostic factors coupled with more accurate pathological and clinical staging of disease and advances in surgical technique. These factors culminate in a more effective treatment of patients at an early stage and at a personalized level. Survival rates in early stage cancer, where the tumour is localized, are extremely good with ~90% of patients experiencing 5-year disease-free survival. Upon spread to localized lymph nodes survival decreases to 50–70% and if distant metastasis has occurred survival is only 12% [1, 2].
Current prognostic staging of CRC
Although multiple CRC subtypes exist, with both molecular and histopathological variances, 90% of CRCs are adenocarcinomas and prognosis is determined through the international tumour, node, metastasis (TNM) staging system alongside the minimal core data set. The TNM staging is based on gross observation and analysis of histopathological tissue sections under the microscope which revolves around the depth of local invasion (T), presence of cancer within the lymph nodes (N) and if the cancer has metastasized (M). TNM staging is excellent at returning prognostic information on a population of patients; however, it is less specialized at predicting prognosis at the level of the individual [3]. A patient’s prognosis is worse the higher the stage they are classified within, however, the TNM system does not differentiate between good and poor outcome of patients within the same stage [4]. There are defined adjuvant treatment guidelines associated with the various stages of CRC [5]. Stage 0 and I cancers will not routinely receive adjuvant chemotherapy and surgical resection is considered curative. Adjuvant therapy is recommended for stage III and IV patients, however, there remains ambiguity about whether to treat all, a subset or no stage II patients with adjuvant chemotherapy [6]. Around 30% of stage II CRC patients will succumb to their disease after surgical resection and, therefore, an accurate and more personalized identification and stratification of high-risk stage II patients, some of whom have comparable or worse outcomes than stage III patients [7], is therefore imperative to increase disease free survival rates. In the UK, pathologists collect a minimal core data set for each patient which helps to identify high-risk stage II CRC cases [8]. Although some parameters within the data set are disregarded in clinical decision making for the management of stage II CRC patients, some features, if present, may invoke the decision to treat the patient: high grade/poor differentiation, pT4 local spread and extramural lymphovascular invasion [9]. However, there is little evidence to date to show the advantage of adjuvant therapy for stage II patients with additional high-risk factors. Furthermore, there are promising histopathological features listed in the literature that have been significantly correlated with poor prognosis but which rarely feature in final clinical reports. There is, for example, growing evidence that immune cell infiltrate and perineural invasion are strongly correlated with poor patient outcome, whereas lymphatic vessel invasion (LVI) and the invasive growth pattern, including tumour budding, are two of the most promising histopathological features that have been significantly associated with lymph node metastasis and disease-specific survival.
Manual reporting yields observer variability
Although histopathological features such as nuclear grade have long been established in the core data sets for CRC prognosis and features such as immune infiltrate, invasive pattern, LVI, lymphatic vessel density (LVD) and the tumour-to-stroma ratio are associated with poor prognosis in the literature, they have also been associated with observer variability in their reporting. Observer variability is an inevitable occurrence when reporting histopathological features by eye under microscopic surveillance; however, it is increased by certain features being obscure under H&E stained tissue with associated retraction artefact, difficult to accurately quantify or if they are rare events. This is particularly true when the calculation of areas in manually determined ‘hot-spots’ is required, such as for LVD and tumour-to-stroma ratio calculations which are both very prone to observer bias and variability. Furthermore, a general consensus on standardized quantification methods is lacking for many of these candidate histopathological features and for this reason they, apart from grading, have not translated into the minimal core data sets of CRC pathological reporting. Although nuclear grading has been reported as a minimal core data set for many years, there have been studies which have also found it to be non-significant [10]. Therefore, there is now a consensus for nuclear grading in CRC to move toward a two-tiered scoring system of ‘well differentiated’ and ‘poorly differentiated’ which eliminates the ‘moderately differentiated’ class and attempts to increase standardization.
Automated image analysis can standardize the quantification of prognostic features
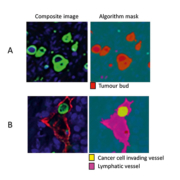
Digital pathology and associated image analysis technology is becoming increasingly sophisticated. It is now possible to create image analysis algorithms that can automatically segment and quantify histopathological features within digital tissue sections with high accuracy. There are multiple advantages to applying image analysis to histopathology reporting that overcome the associated observer variability of manual scoring. Automated image analysis uses standardized algorithms and objectively reports on the features it is programmed to quantify. It does so in a robust manner across all patient samples being tested. Applying image analysis allows a higher degree of accuracy when reporting on the number, area or ratio of specific features across a whole tissue section and so negates the need of manually assigned hot-spots. Furthermore continuous data is captured when image analysis is applied, which allows more accurate clinical cut-offs to be used resulting in a more personalized assessment of a patient’s condition than the more traditional categorical reporting of, e.g. 1+, 2+, 3+ for immunohistochemistry, or ‘well’ or ‘poorly’ differentiated cases. Finally, rare and obscure events that may be missed by the eye are able to be reported with higher confidence when the computer assesses the entire issue section. Jeremy Jass in the late 1980s reported a novel grading system for CRC that included the reporting of the immune infiltrate and the pattern of invasive margin [11]; however, his promising results were not translated into the clinic due to poor reproducibility. Recently, two groups have used image analysis to quantify the immune infiltrate in the form of the immunoscore [12] and the tumour’s infiltrative pattern in the form of tumour budding [13] (Fig. 1A) in a manner that allows standardization, and have shown both features to be prognostically significant. Image analysis has further been used to quantify, amongst others, additional set histopathological features such as for nuclear grading [14], vasculature hot-spots [15] and LVI and LVD (Fig. 1B) [13].
Automated image analysis personalizes CRC prognosis
CRC is a specifically heterogeneous disease and the tumour microenvironment is also a heterogeneous and heterotypic ecology. Therefore, quantifying a single histopathological aspect of a patient’s tumour may not be sufficient for an accurate prognosis across a large population. A recent study by our group used multiplexed immunofluorescence to quantify a suite of histopathological features co-registered across a single CRC tissue section for each patient. Furthermore, the algorithm exported data captured across each nucleus (Fig. 2) in each sample to create a large and complex personalized multi-parametric data set for each patient in the stage II CRC study [16]. A single standardized image analysis algorithm was run across a training set of patients which quantified continuous data from the invasive front of stage II CRC on the number, shape and extent of: tumour buds, poorly differentiated clusters, LVI, LVD, tumour-to-stroma ratio, tumour gland morphology as well as multiple measurements across each nucleus to increase the accuracy of nuclear grading. The resultant ‘big-data’ was distilled through machine learning to identify the optimal parameter set to stratify patients into a high or low risk of disease-specific death. The result was the identification of a novel feature that was independently significant and where the addition of any other measured feature into the model added no further significance to patient stratification. This feature was the mean area of poorly differentiated clusters (area PDC) across the invasive front. The data from the training cohort was validated across a larger independent cohort and again the novel feature held more significance for patient risk stratification in stage II CRC than any of the other more established histopathological feature measured in the study. Furthermore, mathematical modelling was employed to identify if any of the parameters from the clinical pathology report added value to the prediction of disease specific death. By performing this analysis it was found that pT stage and differentiation added further value and were incorporated into a Novel Prognostic Index alongside the area PDC. This novel index outperformed the clinical gold standard of pT staging by almost twofold (Fig. 3).
Big-data and personalized pathology augments clinical staging
The idea behind big-data pathology is to include as much data as possible about each single patient and so to move towards a more personalized prognosis. The acquisition of quantitative data through image analysis and molecular pathology lends itself very well to big-data pathology, where the vast data sets can be mined through sophisticated machine learning algorithms to identify the optimal parameters to answer the clinical question. However, clinical staging and reporting has stood the test of time and it is imperative to include data such as this in any integrative model. As it becomes easier and cheaper to acquire large, reproducible and standardized data sets, modern pathology will become more personalized and patient outcome will improve due to tailored treatment regimens directed at individual patients.
References
1. Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011; 61(4): 212–236.
2. Shah R, Jones E, Vidart V, Kuppen PJ, Conti JA, Francis NK. Biomarkers for early detection of colorectal cancer and polyps: systematic review. Cancer Epidemiol Biomarkers Prev. 2014; 23(9):1712–1728.
3. Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet 2014; 383(9927):1490–1502.
4. Lea D, Haland S, Hagland HR, Soreide K. Accuracy of TNM staging in colorectal cancer: a review of current culprits, the modern role of morphology and stepping-stones for improvements in the molecular era. Scand J Gastroenterol. 2014; 49(10):1153–1163.
5. Poston GJ, Tait D, O’Connell S, Bennett A, Berendse S. Diagnosis and management of colorectal cancer: summary of NICE guidance. BMJ 2011; 343:d6751.
6. Dotan E, Cohen SJ. Challenges in the management of stage II colon cancer. Semin Oncol. 2011; 38(4):511–520.
7. Urquhart R, Bu J, Grunfeld E, Dewar R, MacIntyre M, Porter GA. Examining stage IIB survival in a population-based cohort of patients with colorectal cancer. Cancer 2012; 118(23):5973–5981.
8. Loughrey MB, Quirke P, Shephard NA. Standards and datasets for reporting cancers. Dataset for colorectal cancer histopathology reports July 2014. The Royal College of Pathologists 2014. The cancer datasets are a combination of textual guidance, educational information and reporting proformas to enable consistent grading and staging. (https://www.google.co.uk/?gws_rd=ssl#q=Standards+and+datasets+for+reporting+cancers+Dataset+for+colorectal+cancer+histopathology+reports+July+2014)
9. Morris EJ, Maughan NJ, Forman D, Quirke P. Who to treat with adjuvant therapy in Dukes B/stage II colorectal cancer? The need for high quality pathology. Gut 2007; 56(10):1419–1425.
10. Ratto C, Sofo L, Ippoliti M, Merico M, Doglietto GB, Crucitti F. Prognostic factors in colorectal cancer. Literature review for clinical application. Dis Colon Rectum 1998; 41(8):1033–1049.
11. Jass JR, Love SB, Northover JM. A new prognostic classification of rectal cancer. Lancet 1987; 1(8545):1303–1306.
12. Galon J, Mlecnik B, Bindea G, Angell HK, Berger A, Lagorce C, Lugli A, Zlobec I, Hartmann A, et al. Towards the introduction of the Immunoscore in the classification of malignant tumors. J Pathol. 2014; 232(2):199–209.
13. Caie PD, Turnbull AK, Farrington SM, Oniscu A, Harrison DJ. Quantification of tumour budding, lymphatic vessel density and invasion through image analysis in colorectal cancer. J Transl Med. 2014; 12:156.
14. Rathore S, Hussain M, Aksam Iftikhar M, Jalil A. Novel structural descriptors for automated colon cancer detection and grading. Comput Methods Programs Biomed. 2015; 121(2):92–108.
15. Kather JN, Marx A, Reyes-Aldasoro CC, Schad LR, Zollner FG, Weis CA. Continuous representation of tumor microvessel density and detection of angiogenic hotspots in histological whole-slide images. Oncotarget 2015; 6(22):19163–19176.
16. Caie PD, Zhou Y, Turnbull AK, Oniscu A, Harrison DJ. Novel histopathologic feature identified through image analysis augments stage II colorectal cancer clinical reporting. Oncotarget 2016; doi: 10.18632/oncotarget.10053 [Epub ahead of print].
Acknowledgment
This article is based on the author’s recently published paper: Caie PD, Zhou Y, Turnbull AK, Oniscu A, Harrison DJ. Novel histopathologic feature identified through image analysis augments stage II colorectal cancer clinical reporting. Oncotarget 2016; doi: 10.18632/oncotarget.10053 [16].
The author
Peter Caie PhD
Quantitative and Digital Pathology, School of Medicine, University of St Andrews, North Haugh, St Andrews, UK
*Corresponding author
E-mail: Pdc5@st-andrews.ac.uk