Sepsis: earlier organism identification using MALDI-TOF
Sepsis is a life threatening inflammatory disorder and the immune systems response to infection. It is one of the leading causes of death in hospitalized patients worldwide with 1.8 million cases annually. Improvement in survival remains contingent on early recognition of the causative organism to enable targeted antimicrobial therapy.
by Kelly Marie Ward and Rhian Harris
Sepsis incidence
Sepsis is a life threatening inflammatory disorder and the immune systems response to infection [1]. It is one of the leading causes of death in hospitalized patients worldwide with 1.8 million cases annually [2]. Each year, 37 000 deaths are caused by sepsis in the UK [2–4]. Mortality rates remain between 25–30% for severe sepsis and 40–70% for septic shock, despite advances in pharmacotherapy and supportive care [1] and various campaigns, e.g. the Surviving Sepsis Campaign (SSC) [3]. This is mainly due to poor identification and delayed interventions [3]. Data from the SSC showed a mortality rate of 39.8% among 15 022 patients and 39.8% of those admitted to critical care in England and Wales die in hospital [2]. A hospital admission with severe sepsis places the patient at a level of risk 6–10-fold greater than admission with an acute myocardial infarction and 4–5 times greater than if they had suffered an acute stroke [2].
What is sepsis?
The American College of Chest Physicians and the Society of Critical Care Medicine classified the continuum of an inflammatory response to microorganisms as ‘systemic inflammatory response syndrome’ (SIRS) [1]. SIRS is a collection of signs that show the body is reacting to a range of injuries or illnesses and it is not specific to infection [3]. It is identified when two of the following symptoms – fever, tachycardia, tachypnea and leukopenia are met in the absence of an infection [3].
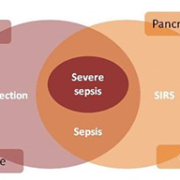
Uncomplicated sepsis is the presence of an infection in association with SIRS [1] in the absence of organ dysfunction [4]. Bacteria that cause infection can enter the body via breaks in the skin, catheters and underlying infections in the urinary, respiratory or gastrointestinal tract [5]. Sepsis can be defined as ‘a systemic disease that is caused by the spread of microorganisms and their toxins via the circulating blood’ [6]. The endo and exotoxins produced by different organisms often lead to an inflammatory response of varying severities [7]. Severe sepsis occurs when SIRS is accompanied by infection and organ dysfunction [4]. Figure 1 taken from Royal College of Physicians: Acute Care Toolkit 9: Sepsis; September 2014 [4] demonstrates this balance.
Pathophysiology of sepsis
The pathophysiology of sepsis involves a complex interaction of proinflammatory and anti-inflammatory mediators in response to pathogen invasion [1]. When an infectious agent invades the host, an innate response is triggered via toll-like receptors (TLR) [8]. These are trans-membrane proteins with the ability to promote signalling pathways downstream and trigger cytokine release, neutrophil activation and stimulation of endothelial cells [8]. The cytokines such as interleukin (IL)-1 and IL-6 are released from the cells where inflammatory reactions have commenced. They stimulate lymphocytes and mononuclear cells to produce further cytokines, resulting in the recruitment and migration of further cells to the site or organ where inflammation is occurring [9]. This leads to endothelium damage, vascular permeability, microvascular dysfunction, coagulation pathway activation and impaired tissue oxygenation resulting in the cascade of sepsis [1]. There is activation of humoural and cell-mediated immunity with specific B and T cell responses and both pro and anti-inflammatory cytokine release [8]. Adaptive immunity is triggered and the inflammatory cascade of sepsis occurs where the balance is shifted towards cell death and a state of relative immunosuppression and end organ dysfunction ensues with hemodynamic changes causing elevated cardiac output and generalized vasodilation described as shock [8]. As the inflammatory response progresses, myocardial depression is more pronounced resulting in a falling cardiac output. There is capillary leak and pulmonary edema that may progress to acute lung injury. Renal failure then follows accompanied by alterations in the coagulation cascade towards a pro-coagulant and antifibrinolytic state. The development of ‘disseminated intravascular coagulation’ (DIC) in severe sepsis is a predictor of death and the development of multiorgan failure [8]. The respiratory, genitourinary and gastrointestinal systems are most commonly infected and pneumonia is the most common presentation leading to sepsis [1].
Clinical presentation and diagnosis
There are a variety of symptoms that can indicate sepsis, including fever, chills, decreased blood pressure, shaking, skin rash, confusion and a rapid heartbeat [10].The clinical diagnosis of sepsis is most often made before culture results are available and although localized signs and symptoms may be present, organ hypoperfusion or shock can occur without the knowledge of the cause [1]. Fever is the most common manifestation of sepsis and 40% of those patients will have hypotension [1].
A vast array of laboratory tests are required for the diagnosis and management of sepsis including full blood counts, basic metabolic panels, lactate and liver enzyme levels and C-reactive protein. In the Microbiology laboratory we would expect to receive, blood cultures: two peripheral and from each indwelling catheter, urine, stools if symptoms of diarrhoea, sputum and skin and soft tissue for culture if clinically significant [1] Currently blood cultures are the definitive diagnosis tool when septicemia is suspected [11]. Blood culture systems have evolved over time to ensure optimum isolation of any organisms present by adding different nutrients, introduction of automated systems and increasing the detection rate of a positive as a result of new software [12]. They are used to detect the presence of any microorganisms present by providing optimum conditions for growth. However in 50–65% of patients the blood culture is often negative [1].
Early management and identification of infectious cause
Due to the high mortality rates the early identification and management of sepsis is crucial and requires respiratory stabilization followed by fluid resuscitation, vasopressor therapy, infection identification and control and prompt antibiotic administration [1].
The SSC published ‘The Resuscitation Bundle’ which comprised a set of tasks to be to be completed within the first 6 hours after the clinical identification of sepsis [2]. The first four tasks were:
- Measure serum lactate
- Obtain blood cultures prior to antibiotic administration
- Broad-spectrum antibiotics to be given within 1 hour of presentation
- Source of infection to be identified and drained within 6 hours.
These tasks involve the Pathology laboratory and it was established that systems within healthcare environments needed to be well designed and implemented to ensure that the appropriate investigations, equipment and treatments were available at the point of care [2].
The length of time that it takes for the correct identification of the causative organism has many ramifications both clinically and financially. Empiric antibiotic therapy is based on the most likely source, clinical context, recent antibiotic use and local resistance patterns. This should be narrowed when the causative agent has been identified to reduce the risk of resistance or superinfection [1]. The length of time the patient is prescribed such antibiotics may be reduced if the causative organism is characterized sooner, and inappropriate therapy changed accordingly. The use of targeted narrow spectrum antibiotics might reduce bed days resulting in a financial saving. Using antibiotics more than is required in both humans and animals has resulted in the increased emergence of antibiotic resistance [13]. New antibiotics are produced very slowly and as a result it is important to limit any likelihood of the spread of resistance between organisms by only prescribing the antibiotic necessary [14]. Also early appropriate antibiotic therapy is associated with improved clinical outcomes [1]. The antibiotics should be administered within 1 hour of suspected sepsis. In septic shock early antibiotic therapy increases survival and for each hour this is delayed the survival rates decrease by 8% [1]. The possible benefits to the patient in terms of improved treatment of infections, the potential to reduce costs and an attempt to reduce the emergence of antibiotic resistance have led to the development and research into more rapid diagnosis techniques.
MALDI-TOF and the Bruker Sepsityper blood culture kit
Matrix-assisted laser desorption/ionization–time of flight (MALDI-TOF) spectroscopy can identify organisms from intact cells based on the profile of different proteins and relative molecular mass [15]. A smear of the cultured organism is placed onto a stainless steel target plate, with matrix placed over the top. Matrix is used as it prevents fragmentation of higher mass molecules. The laser is fired at the smear generating a cloud of ions which are accelerated up the flight tube to the detector, where the time of flight is converted to Daltons (Da)/molecular mass. The heavier the molecular mass of the ion the greater the time of flight (Fig. 2).
This is also known as proteomic profiling. A spectrum is produced for each organism based on their mass/charge ratio, which is determined by the different molecular mass and charge of the ions present for the organism in question [15]. A spectrum is produced with a variety of peaks each one representing a different molecular fragment which has been released as a result of the laser desorption [15]. This spectrum is then compared to the database for possible matches and is scored based on the number of peaks that match the corresponding organism (Fig. 3).
This method can be used to identify bacteria, yeasts, moulds, mycobacteria and Nocardia to species level using species-specific spectral patterns [16]. Using this method, identification from a bacterial culture can be achieved in 30–60 seconds.
MALDI-TOF spectroscopy can also be used to identify organisms directly from blood culture bottles using the Sepsityper kit extraction method (Fig. 4). This takes approximately 30 minutes and allows accurate identification to species level on day 1 of the bottle being flagged as positive. The use of the Sepsityper kit could enhance task three of the SSC Resuscitation bundle by allowing earlier targeted therapy. Components within the blood culture such as red cells, white blood cells and serum can interfere with the analysis resulting in the formation of additional spectral peaks [17]. These peaks will not be found in the database and will result in difficulty in interpreting the results, which is why the extraction kit by Bruker has been developed. The development of this kit has allowed purification and extraction to be carried out to optimize recovery of the bacteria present in the blood culture sooner. This is carried out by a series of centrifugation steps to separate any organisms present from the blood and fluid present in the blood culture and also formic acid to breakdown the cell wall of the organism to aid identification using MALDI-TOF technology (Bruker – Introduction for use Maldi Sepsityper kit (Accessed 2015).
Our laboratory evaluated the use of the Sepsityper kit to identify the causative organism direct from the positive blood culture bottle using MALDI-TOF spectroscopy. The results were retrospectively analysed to determine if there would have been a change to the antibiotic therapy if this method was in routine use.
Study results
Two hundred and thirty-six positive blood cultures were analysed retrospectively and compared against current laboratory methods. The results are shown in Table 1.
Table 1 shows the percentage of successful identifications achieved by using the Bruker Sepsityper method. The percentage of blood cultures that achieved successful identification within 1 hour of becoming positive was 75.42% (green plus yellow rows). A score of above 1.8 indicates a secure genus and probable species identification (green row) [18], a score between 1.6 and 1.8 indicates probable species identification (yellow row) [18]. Any score below 1.6 cannot be accepted as a reliable identification (red row). There was a 93.33% agreement of identification between the Bruker Sepsityper kit or direct MALDI-TOF identification versus the BD Phoenix and other conventional laboratory methods.
The previous antibiotic treatment, the clinical history of the patient and the identification of the organism produced by the Bruker Sepsityper kit on day one was analysed retrospectively by the consultant microbiologist to determine if there would have been any clinical impact if the identification of the organism had been known on day 1. As the organism that is causing the infection is not known immediately, patients are started on broad-spectrum or a combination of antibiotics when bacteremia is suspected; however, incorrect or insufficient therapy has been associated with increased mortality, morbidity and increased hospital stay [19]. The consultant microbiologist determined that 26 (11%) out of the 236 blood cultures analysed would have indicated a requirement for the patient to have their antibiotic therapy altered in some way. Sixteen of the 26 positive blood cultures indicated that the patients’ antibiotic therapy could be reduced from a broad-spectrum antibiotic to a narrower spectrum antibiotic. This can have huge cost savings implications as well as reduce the likelihood of resistance emerging against broad-spectrum antibiotics [20]. Knowing the identification of the organism on the day the blood culture bottle is flagged as positive enables the antimicrobial therapy to be changed accordingly therefore helping to reduce the emergence of resistance, provide targeted therapy for better treatment outcomes and reduce bed days spent in hospital. Improvement in survival remains contingent on the early recognition and management of severe sepsis and septic shock [1].
References
1. Gauer RL. Early Recognition and Management of Sepsis in Adults: The First Six Hours. Am Fam Physician. 2013; 88: 44–53.
2. Daniels R. Surviving the First Hours In Sepsis: getting the basics right (an intensivists perspective). J Antimicrob Chemother. 2011; 66(Suppl 2): ii11–23.
3. McClelland H, Moxon A. Early identification and treatment of sepsis. Nursing Times 2014; 110: 14–17. (http://www.nursingtimes.net/Journals/2014/01/17/q/v/z/220114-Early-identification-and-treatment-of-sepsis.pdf)
4. Royal College of Physicians. Acute Care Toolkit 9: Sepsis; September 2014. (https://www.rcplondon.ac.uk/sites/default/files/acute_care_toolkit_9_sepsis.pdf)
5. Public Health England. Investigation of blood cultures. Bacteriology 2014; B37(8). (https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/372070/B_37i8.pdf)
6. Odeh M. Sepsis, septicaemia, sepsis syndrome, and septic shock: the correct definition and use. Postgrad Med J. 1996; 72(844): 66.
7. Martin GS. Sepsis, severe sepsis and septic shock: changes in incidence, pathogens and outcomes. Expert Rev Anti-infect Ther. 2012; 10(6): 701–706.
8. Ventetuolo CE, Levy MM. Sepsis: a clinical update. Clin J Am Soc Nephrol. 2008; 3: 571–577.
9. Roitt I, Brostoff J, Male D. Cell migration and inflammation. In: Cook L, Immunology, 4th ed. Mosby 1998.
10. Severe sepsis/septic shock, recognition and treatment protocols. Stony Brock Medicine 2013. (http://www.survivingsepsis.org/sitecollectiondocuments/protocols-sepsis-treatment-stony-brook.pdf)
11. Previsdomini M, Gini M, et al. Predictors of positive blood cultures in critically ill patients: a retrospective evaluation. Croat Med J. 2012; 53(1): 30–39.
12. Zadroga R, Williams DN, et al. Comparison of 2 blood culture media shows significant differences in bacterial recovery for patients on antimicrobial therapy. Clin Infect Dis. 2012; 56(6): 790–797.
13. Rao GG. Risk factors for the spread of antibiotic-resistant bacteria. Drugs 1998; 55(3): 323–330.
14. Guidos RJ. Combating antimicrobial resistance: policy recommendations to save lives. Clin Infect Dis. 2011; 52(5): 397–428.
15. Carbonnelle E, Beretti JL, et al. (2007). Rapid identification of Staphylococci isolated in clinical microbiology laboratories by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2007; 45(7): 2156–2161.
16. Stevenson LG, Drake SK, Murray PR. Rapid identification of bacteria in positive blood culture broths by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2010; 48(2): 444–447.
17. Lagacé-Wiens PRS, Adam HJ, et al. Identification of blood culture isolates directly from positive blood cultures by use of matrix-assisted laser desorption ionization–time of flight mass spectrometry and a commercial extraction system. J Clin Microbiol. 2012; 50(10), 3324–3328.
18. El-Bouri K, Johnston S, et al. Comparison of bacterial identification by MALDI-TOF mass spectrometry and conventional diagnostic microbiology methods: agreement, speed and cost implications. Br J Biomed Sci 2012; 69(2): 47–55.
19. Kollef MH. Broad-spectrum antimicrobials and the treatment of serious bacterial infections: getting it right up front. Clin Infect Dis. 2008; 47(1): 3–13.
20. Rüttimann S, Keck B, et al. Long-term antibiotic cost savings from a comprehensive intervention program in a medical department of a university-affiliated teaching hospital. Clin Infect Dis. 2004; 38(3): 348–356.
The authors
Kelly Marie Ward* MSc, FIBMS; Rhian Harris MSc, AIBMS
Royal Glamorgan Hospital Microbiology Laboratory, Cwm Taf University Health Board, Llantrisant, Glamorgan, UK
*Corresponding author
E-mail: Kelly.Ward@wales.nhs.uk