LC-MS/MS-based H-type determination of Escherichia coli
A comparative analysis of serotyping and mass spectrometry (MS) methods for the determination of the flagellar type (H type) of clinically isolated Escherichia coli has been performed. In this analysis, it was shown that determination of the correct H type of a clinical E. coli strain was better achieved by MS than serotyping. Whole genome sequencing was used for the validation of this analysis.
by M. Chan, Dr H. Chui, D. Hernandez, Dr G. Wang and Dr K. Cheng
Background
During outbreaks of disease caused by pathogenic Escherichia coli, it is important to be able to identify the precise E. coli strain involved in order to track and prevent the spread of infection. Typically, the identification of E. coli strains has been based on the serotype of the cell surface antigens such as the lipopolysaccharide O antigen and the flagellar H antigen. Even though serotyping is viewed as the ‘gold standard’ for O- and H-type determination, this technique does have its downfalls. These conventional serotyping methods are based on antisera, which makes procedures costly and laborious to perform because of the variable quality of antibody preparations and the number of antibody agglutination reactions needed to assign a final classification [1, 2, 3]. Also, when bacterial cells do not generate lipopolysaccharide on the surface, the cultured colonies become ‘rough strains’. This makes both O- and H-antigen identification by antibody-based agglutination problematic despite the cellular motility and presence of the flagella H-antigen structure [1, 3, 4]. In addition, flagellar serotyping needs the induction of motility, which can take up to two weeks, and so does not result in the fast identification required in an E. coli outbreak situation [5].
A promising technique for E. coli H-type identification is the use of a liquid chromatography-tandem mass spectrometry (LC-MS/MS) method, termed ‘MS-H’. In our own analytical assays, we harvest, enrich and digest the flagella proteins. LC-MS/MS uses liquid chromatography to separate flagellar fragments after trypsin digestion, and mass spectrometry to analyse and determine the protein sequences of these fragments. The MS results are then analysed against a curated database of H antigen variants to determine the H type. Compared with serotyping, MS-H has a high throughput, requires less labour, needs less time to perform, and provides sequence-level information.
Comparative H typing of E. coli with serotyping and MS-H
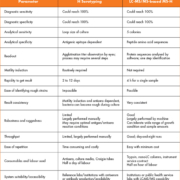
Using reference strains of all the different forms of E. coli flagella, it was shown that all 53 different types of H antigen can be determined through MS-H. Table 1 details the comparisons between H serotyping and MS-H [1]. It is important to note that both methods can reach 100% sensitivity and specificity for H-type determination. However, MS-H also provides sequence-level identification of the H type. This is important because this provides more reliable results, while antisera serotyping only provides a visual conformation, making there more of a chance for subjective and inaccurate determination of the H type.
Flagella motility induction is commonly used in serotyping. This increases the time-length of the procedure and requires more hands-on involvement [5]. In comparison, MS-H uses motility induction much less frequently, which makes MS-H identification of E. coli H type quicker and less labour-intensive.
Clinically isolated strains of E. coli that had already had their H types determined by serotyping were supplied from three provinces within Canada. A preliminary H-type analysis of the same E. coli isolates was performed by MS-H [5]. The workflow that was completed to analyse the H type of these strains is detailed in Figure 1.
Things to note within the workflow are that vortexing the suspended culture in Step 2 was to shear the flagella away from the E. coli. Also in Step 2, the flagella were trapped onto a filter membrane. This serves as an isolation step to separate the flagella from the supernatant as well as an enrichment process to concentrate the flagella. We then compared the flagellar sequence obtained by MS to a curated database formed from NCBI flagella protein sequence entries. A curated database was necessary because a public database can be problematic because of size, specificity and inconsistent annotation of the data for the specific flagellar protein entries. The curated database focuses on the flagellar proteins with fit-for-purpose annotations, thus allowing differentiation between the 53 different H types better than the public database [6].
On comparison of the H types identified by serotyping and MS-H, the majority of the results agreed with one another. However, there were some discrepancies between the two methods. In these cases, whole genome sequencing and polymerase chain reaction (PCR) detection was used to identify the correct H type [5] and these results predominantly agreed with those obtained by MS-H.
Even though PCR as a form of detection for the flagella gene is popular, the accuracy for determining all forms of flagella types through PCR is low and the use a many different primers had to be implemented, making it not ideal for the detection of an unknown H type [5]. Whereas whole genome sequencing may be more expensive then PCR detection, it was much more accurate at determining the correct H-type allele than PCR detection. Thus moving into the confirmational assays for the comparative analysis of MS-H determination and H serotyping, whole genome sequencing was only used to resolve disputed results between the two methods as it is not exclusively representative of a host’s flagella phenotype. On the other hand serotyping and MS-H is representative of the bacteria’s H type. Whole genome sequencing isn’t ideal for identifying clinical strains of E. coli for it is laborious and a long process compared to MS-H which is faster and contains less labour-intensive.
MS-H in clinical laboratories
Having shown that MS-H determined E. coli H type with high accuracy and in a short time frame, the application of H typing through MS-H could become very useful for the identification of unknown E. coli strains in a clinical laboratory. This is especially useful in the identification of an E. coli strain during a disease outbreak [7]. Faster identification of the pathogenic strain of E. coli would in turn also help combat the pathogenic E. coli more quickly.
Without the use of antibodies for agglutination and procedures for motility induction that are required in serotyping, MS-H is much less laborious in comparison. This would greatly benefit the professionals in the medical microbiology and public health laboratories who perform H typing.
Advantages and disadvantages of MS-H
It is significant that MS-H better determined the H type compared to conventional serotyping. As MS-H is faster, less laborious, provides sequence-level identification and has a higher throughput than serotyping, MS-H can be very useful in rapid and accurate identification of E. coli flagellar antigens. This may be a little over-idealistic as LC-MS/MS machines are very expensive and serotyping has been around for a very long time. It might be difficult currently for clinical laboratories to afford a LC-MS/MS machine or to change their workflow to incorporate MS-H as their method of H determination. However, mass spectrometer platforms are becoming more common in clinical laboratories. The uses of matrix-assisted laser desorption/ionization (MALDI) mass spectrometers have shown varying successes in microbiological identification. Also the use of MS isn’t just limited to H typing. MS could extend to the determination of the lipopolysaccharides (O antigen) of E. coli, toxins, and other relevant molecules within the spectrum of the mass spectrometer. This would not only determine the surface antigens of pathogenic E. coli, it would also give a broader profile of a pathogenic E. coli.
Conclusion and future directions
MS has potential to determine the H antigen type of E. coli better than serotyping, as shown in a comparative study between MS-H and serotyping through the use of a mass LC-MS/MS platform. MS-H provides sequence level identification of the flagellum type, whereas serotyping only provides visual agglutination assays for positive results and is therefore more prone to errors such as false positives and misidentification. Also, without the need for antisera, MS-H is less laborious, requires less time, and has a high throughput.
With the completion of the preliminary assay results of the comparative study between MS-H and serotyping in the determination of the flagella type, we are currently working on a country-wide thorough validation of the platform.
References
1. Cheng K, Drebot M, McCrea J, Peterson L, Lee D, McCorrister S, Nickel R, Gerbasi A, Sloan A, Janella D, Van Domselaar G, Beniac D, Booth T, Chui L, Tabor H, Westmacott G, Gilmour M, Wang G. MS-H: A novel proteomic approach to isolate and type the E. Coli H antigen using membrane filtration and liquid chromatography-tandem mass spectrometry (LC-MS/MS). PLoS One 2013; 8(2): 1–12.
2. Tenover FC, Arbeit RD, Goering RV. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: A review for healthcare epidemiologists. Molecular Typing Working Group of the Society for Healthcare Epidemiology of America. Infect Control Hosp Epidemiol. 1997; 18(6): 426–439.
3. Machado J, Grimont F, Grimont PA. Identification of Escherichia coli flagellar types by restriction of the amplified fliC gene. Res Microbiol 2000; 151(7): 535–546.
4. Edwards PR, Ewing WH. Edwards and Ewing’s Identification of Enterobacteriaceae, p536. Elsevier 1986. ISBN 978-0444009817.
5. Cheng K, Sloan A, Peterson L, McCorrister S, Robinson A, Walker M, Drew T, McCrea J, Chui L, Wylie J, Bekal S, Reimer A, Westmacott G, Drebot M, Nadon C. Knox D, Wang G. Comparative study of traditional flagellum serotyping and liquid chromatography-tandem mass spectrometry-based flagellum typing with clinical Escherichia coli isolates. J Clin Microbiol. 2014; 52(6): 2275-2278.
6. Cheng K, Sloan A, McCorrister S, Babiuk S, Bowden TR, Wang G, Knox D. Fit-for-purpose curated database application in mass spectrometry-based targeted protein identification and validation. BMC Res Notes 2014; 7: 444.
7. Cheng K, Sloan A, McCorrister S, Peterson L, Chui H, Drebot M, Nadon Celine, Knox D, Wang G. Quality evaluation of LC-MS/MS-based E. coli H antigen typing (MS-H) through label-free quantitative data analysis in a clinical sample setup. Proteomics Clin Appl. 2014; 8: 963–970.
The authors
Michael Chan*1,2, Huixia Chui1,3 MD, Drexler Hernandez1,2, Gehua Wang*1 MD and Keding Cheng*1,4 MD
1National Microbiology Laboratory, Public Health Agency of Canada, Winnipeg, MB, Canada
2University of Manitoba, Winipeg, MB, Canada
3Centre of Disease Control and Prevention, Henan Province, PR China
4Department of Human Anatomy and Cell Sciences, Faculty of Medicine, University of Manitoba, Winnipeg, MB, Canada
*Corresponding authors
E-mail: M. Chan, umchanm@myumanitoba.ca; G. Wang, gehua.wang@phac-aspc.gc.ca; K. Cheng, keding.cheng@phac-aspc.gc.ca.