A new approach for diagnosis of Clostridium difficile based on evolution of a unique volatile organic compound identifier
Clostridium difficile is a major cause of nosocomial infections and rapid diagnosis of the disease is essential for infection control. Several methods for C. difficile detection are employed in clinical laboratories; each method has its advantages and disadvantages. A novel method has recently been developed that allows differentiation between C. difficile-positive and -negative stool samples based on volatile organic compound evolution and their detection by headspace solid-phase microextraction gas chromatography–mass spectrometry.
by Dr Emma Tait, Prof. Stephen P. Stanforth, Prof John D. Perry and Prof. John R. Dean
Introduction
Clostridium difficile is a Gram-positive anaerobe and the causative agent of C. difficile infection (CDI). CDI is a major healthcare problem with a total of 14,687 cases reported in patients aged 2 years and over in England between April 2012 and March 2013 [1]. C. difficile is a spore-forming bacterium; dormant spores are resistant to antibiotics, heating and chemicals such as disinfectants and, therefore, can persist on surfaces and survive for long periods in the environment [2]. Ingestion of spores and their subsequent germination in the gut allows the proliferation of C. difficile in patients whose normal gut flora has been severely reduced following antibiotic treatment. Following germination, vegetative C. difficile can produce toxins and is susceptible to antibiotic treatment. Pathogenic C. difficile releases two types of toxins, toxin A and toxin B, and it is these toxins that cause the symptoms associated with CDI [3]. Clinical symptoms of C. difficile infection (CDI) include mild to severe diarrhoea, which can lead to pseudomembranous colitis and death. Diagnosis of CDI includes both clinical manifestations of symptoms supported by laboratory findings.
Diagnosis of C. difficile infection
Rapid diagnosis of CDI is essential to allow the most appropriate treatment to be prescribed, to enable proper use of hospital isolation facilities and to reduce the spread of the infection. Routine diagnostic methods in clinical microbiology laboratories employ a variety of techniques for diagnosis of CDI. These include immunoassays for detection of glutamate dehydrogenase (GDH) antigen and toxins, polymerase chain reaction (PCR) for detection of toxin B or isolation of C. difficile by culture (followed by confirmation of toxigenicity, e.g. using PCR). Immunoassays and PCR methods are typically highly automated and deliver rapid results and these have largely replaced the traditional cell cytotoxicity assay, which requires propagation of cell lines and takes days rather than hours to provide results.
Toxin immunoassays, although relatively inexpensive with rapid turnaround time, have limitations in terms of their sensitivity and specificity leading to false-negative results and false-positive results [4]. Immunoassays for GDH are typically highly sensitive for detection of C. difficile but lack specificity. Culture media for the isolation of C. difficile typically incorporate the antibiotics D-cycloserine and cefoxitin which suppress commensal bacteria; such media are based on the formulation recommended by George et al. [5]. Isolation of C. difficile via culture is sensitive but can take several days to obtain results and there may be a heavy growth of other fecal bacteria, particularly when reduced antibiotic concentrations are used [6]. Recent developments in C. difficile detection include using a chromogenic substrate which is structurally similar to naturally occurring substrate used by C. difficile toxins [7]. This allows the detection of toxigenic C. difficile, i.e. only pathogenic strains are targeted.
Identification of C. difficile-positive stool samples using gas chromatography has previously been explored [8]; volatile organic compounds (VOCs) such as p-cresol and short chain fatty acids were identified as potential markers for C. difficile. However, these methods suffered from a lack of specificity, particularly due to the high number of false positives obtained following the detection of p-cresol and isocaproic acid in stool samples without C. difficile. As a consequence, gas chromatography methods were deemed unsuitable for C. difficile detection [8]. More recent attempts to use bacterial VOC analysis as a tool for C. difficile identification have used headspace solid-phase microextraction (HS-SPME) as a VOC collection method coupled with gas chromatography–mass spectrometry (GC-MS) for VOC separation and detection [9].
Development of a novel method for detection of C. difficile in stools
A novel method for rapid identification of C. difficile in stool samples has been developed using the analysis of VOCs. Use of synthetic enzyme substrates is an effective means of differentiating bacteria, for example in chromogenic culture media [10]. These types of culture media incorporate chromogenic enzyme substrates where the action of a specific enzyme on the substrate liberates a molecule that is detectable visually, allowing the detection of pathogenic bacteria [10]. The philosophy behind the use of substrates in culture media can be applied to the analysis of bacterial VOCs, where a substrate is incorporated into a clinical sample inoculated in liquid media; the cleaved product is volatile and detectable using an analytical method such as HS-SPME-GC-MS. The detection of VOCs liberated following enzyme activity increases the specificity of bacterial VOC profiles, as these liberated VOCs act as markers for a particular species, hence aiding identification of bacteria. This approach was applied to the detection of C. difficile in stool samples.
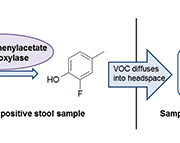
p-Cresol is formed in C. difficile by the decarboxylation of p-hydroxyphenylacetic acid. The enzyme responsible for this decarboxylation is p-hydroxyphenylacetate decarboxylase. It has been established that the hydroxyl group in the para position on the phenyl ring is an essential requirement for decarboxylation to occur [11]. C. difficile is almost unique in its ability to form p-cresol using this pathway, with the exception of a Lactobacillus strain [12]. This was exploited in the development of the novel method that allows successful differentiation between C. difficile culture-positive and -negative stool samples based on VOC generation from an enzyme substrate [13]. 3-Fluoro-4-hydroxyphenylacetic acid was used as a substrate for p-hydroxyphenylacetate decarboxylase; the evolution of the VOC 2-fluoro-4-methylphenol indicated the presence of C. difficile (Fig. 1). VOCs were detected using HS-SPME-GC-MS.
The lack of specificity of previous GC methods was often due to the detection of VOCs in C. difficile culture-negative stool samples as these VOCs were generated by commensal bacteria. Techniques employed to reduce background flora, and therefore improve the selectivity and sensitivity of methods, include alcohol shock [14] and the inclusion of antibiotics [5]. The antibiotics D-cycloserine, cefoxitin and amphotericin were added to the sample matrix and an alcohol-shock step was included to suppress background flora present in stool samples. Alcohol shock kills vegetative cells but does not affect the viability of spores and incorporation of sodium taurocholate in a culture medium can subsequently aid germination of spores [6, 15]. Inoculation of stool samples into such a culture medium allows bacterial growth and concomitant generation of VOCs.
The method was tested with 100 stool samples, of which 77 were C. difficile culture-positive and 23 culture-negative. The generation of 2-fluoro-4-methylphenol indicated the presence of C. difficile after overnight incubation. Method specificity and sensitivity were 100 % and 83.1 %, respectively, using 2-fluoro-4-methylphenol as a marker for C. difficile identification (Table 1). The VOCs isocaproic acid and p-cresol were useful indicators for C. difficile-positive stool samples, although were insufficient for identification purposes. Both VOCs, particularly p-cresol, were generated by C. difficile-negative samples; this is in agreement with previous studies [8].
Advantages and disadvantages of VOC method
The method allows the detection of C. difficile with a very high specificity (100%), i.e. 2-fluoro-4-methylphenol was not generated by C. difficile culture-negative stool samples tested. Rapid detection of VOCs was possible with confirmation of the presence of C. difficile within 18 hours. This indicates that the method could be used to screen for C. difficile in stools allowing the prompt diagnosis of culture-positive samples by the detection of 2-fluoro-4-methylphenol. However, a study on method sensitivity in terms of the number of bacterial cells required to generate a positive signal confirmed that identification of C. difficile was possible provided the stool sample contained at least 150 colony forming units (CFU). It is entirely possible that some stool samples will contain much fewer CFU and therefore 2-fluoro-4-methylphenol would not be detected and a false-negative result would be obtained. This limitation is reflected in the method sensitivity (83.1%) after evaluation with 100 stool samples. The method targets all strains of C. difficile and further testing would be required (e.g. using PCR or immunoassay) to distinguish whether positive stool samples contain toxigenic strains. As a result, it is recommended that VOC analysis should be used alongside conventional methods for C. difficile detection, including toxin detection methods, which would allow any false negative results to be eliminated.
Conclusion
C. difficile is a common cause of nosocomial infections and therefore rapid, accurate diagnosis of CDI is of extreme importance for infection control and patient care. There are currently a number of methods used in hospital laboratories for the diagnosis of CDI; however, each method has its drawbacks. A novel approach has been developed for the identification of C. difficile in stool samples that involves the incubation of stool samples in the presence of 3-fluoro-4-hydroxyphenylacetic acid which acts as a substrate for the enzyme p-hydroxyphenylacetate decarboxylase. The success of this new approach is evaluated by its application to 100 stool samples and its ability to differentiate between C. difficile culture-positive and -negative stool samples. It is envisaged that the identification of C. difficile culture-positive stool samples by the analysis of VOCs could allow rapid diagnosis of CDI. In addition, the novel approach of using enzyme substrates that release VOCs that are not normally generated by bacteria, for example fluorinated VOCs, may find application in the identification of other bacterial pathogens in clinical microbiology.
References
1. Public Health England. Summary Points on Clostridium difficile Infection (CDI). 2013; http://www.hpa.org.uk/webc/HPAwebFile/HPAweb_C/1278944283388.
2. Kuijper EJ, Coignard B, Tull P. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin Microbiol Infect. 2006; 12(Suppl 6): S2−S18.
3. Voth DE, Ballard JD. Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev. 2005; 18: 247–263.
4. Eastwood K, Else P, Charlett A, Wilcox M. Comparison of nine commercially available Clostridium difficile toxin detection assays, a real-time PCR assay for C. difficile tcdB, and a glutamate dehydrogenase detection assay to cytotoxin testing and cytotoxigenic culture methods. J Clin Microbiol. 2009; 47: 3211–3217.
5. George WL, Sutter VL, Citron D, Finegold SM. Selective and differential medium for isolation of Clostridium difficile. J Clin Microbiol. 1979; 9: 214–219.
6. Nerandzic MM, Donskey CJ. Effective and reduced-cost modified selective medium for isolation of Clostridium difficile. J Clin Microbiol. 2009; 47: 397–400.
7. Darkoh C, Kaplan HB, DuPont HL. Harnessing the glucosyltransferase activities of Clostridium difficile for functional studies of toxins A and B. J Clin Microbiol. 2011; 49: 2933–2941.
8. Levett PN. Detection of Clostridium difficile in faeces by direct gas liquid chromatography. J Clin Pathol. 1987; 37: 117–119.
9. Garner CE, Smith S, Costello BL, White P, Spencer R, Probert CSJ, Ratcliffe NM. Volatile organic compounds from feces and their potential for diagnosis of gastrointestinal disease. Faseb J. 2007; 21: 1675–1688.
10. Orenga S, James AL, Manafi M, Perry JD, Pincus DH. Enzymatic substrates in microbiology. J Microbiol Meth. 2009; 79: 139–155.
11. Selmer T, Andrei PI. p-Hydroxyphenylacetate decarboxylase from Clostridium difficile. A novel glycyl radical enzyme catalysing the formation of p-cresol. Eur J Biochem. 2001; 268: 1363–1372.
12. Yokoyama MT, Carlson JR. Production of skatole and para-cresol by a rumen Lactobacillus sp. Appl Environ Microbiol. 1981; 41; 71–76.
13. Tait E, Hill KA, Perry JD, Stanforth SP, Dean JR. Development of a novel method for detection of Clostridium difficile using HS-SPME-GC-MS. J Appl Microbiol. DOI: 10.1111/jam.12418.
14. Clabots CR, Gerding SJ, Olson MM, Peterson LR, Gerding DN. Detection of asymptomatic Clostridium difficile carriage by an alcohol shock procedure. J Clin Microbiol. 1989; 27: 3286–3287.
15. Wilson KH, Kennedy MJ, Fekety FR. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J Clin Microbiol. 1982; 15; 443–446.
The authors
Emma Tait1 PhD, Stephen P. Stanforth1 PhD, John D. Perry2 PhD and John R. Dean1* DSc, PhD
1Faculty of Health & Life Sciences, Department of Applied Sciences, Northumbria University, Newcastle-upon-Tyne, UK
2Department of Microbiology, Freeman Hospital, Newcastle-upon-Tyne, UK
*Corresponding author
E-mail: John.dean@northumbria.ac.uk