Improved diagnostics of Tropheryma whipplei
Whipple´s disease, a systemic and ultimately fatal infection with Tropheryma whipplei, is usually easily treated if diagnosed early enough. A novel real-time PCR protocol and fluorescence in situ hybridization provide an improved diagnosis. All results, however, need careful interpretation. We recommend involving specialized centres in the initial diagnosis and patient follow-up.
by Alexandra Wießner, Dr Annette Moter and Dr Judith Kikhney
Whipple’s disease: a fatal infectious disease
Tropheryma whipplei causes a rare, but fatal, bacterial infection: Whipple’s disease. This systemic disease can usually be cured by antibiotic therapy if detected early enough. The key challenge for physicians and microbiologists is to recognize the bacterial origin of various clinical symptoms in time. Diagnosis is still not trivial owing to the rarity of the disease, diverse and unspecific clinical symptoms, the fastidious nature of T. whipplei and the absence of non-invasive serological tests [1]. Improved diagnostic assays for the detection of T. whipplei are very valuable in combination with expertise to interpret the results for fast initiation of treatment.
T. whipplei belongs to the Gram-positive class of Actinobacteria and can be detected intracellularly in vacuoles or as extracellular bacteria in the tissue [2]. It is a slender rod shape, readily visible with Periodic Acid–Schiff (PAS) staining. T. whipplei strains can be cultured in an axenic culture medium supplemented with amino acids, but the slow growth rate means that culture is not an option for routine diagnosis of T. whipplei infection.
Symptoms of Whipple’s disease
In classical Whipple’s disease patients suffer from chronic diarrhoea, weight loss and fever. Molecular methods have detected isolated or systemic T. whipplei infection in almost every organ [joints, central nervous system (CNS), heart valves, skin, eye, lymph node, bone and lung], even in the absence of intestinal involvement. Depending on the location of infection, the symptoms may vary substantially. Often, the diagnosis of Whipple’s disease is delayed as the result of misdiagnosis as sero-negative rheumatoid arthritis, culture-negative endocarditis or neurological disorders. The involvement of the CNS is especially dramatic, as damage caused by the bacteria is often irreversible and antibiotic treatment may no longer be effective enough to cure the infection [3].
Transmission and asymptomatic carriage
To complicate the picture even more T. whipplei has been found in healthy carriers at an estimated prevalence in the population of <1–4% [4, 5]. This means that the detection of T. whipplei in stool or saliva may not be indicative of Whipple’s disease and in this case does not necessarily require antibiotic treatment. A higher prevalence has been found in high risk populations for direct or indirect faecal–oral transmission, such as sewage workers [5], homeless people and family members of Whipple´s disease patients. As T. whipplei is common in the environment, it is assumed that Whipple’s disease patients must have an immunological predisposition for developing a chronic infection instead of being only transiently colonized [6].
The current transmission model assumes that T. whipplei is taken up orally, probably in early childhood, leading to temporary asymptomatic carriage, self-limiting gastroenteritis, fever, or cough [1, 7, 8]. In most cases a protective humoral and cellular immune response prevents T. whipplei infection. However, in predisposed persons T. whipplei may spread systemically over the years resulting in Whipple´s disease.
Diagnosis of Whipple’s disease
Currently, Whipple’s disease is most often detected through PAS staining of biopsies from the lower duodenum or jejunum showing PAS-positive macrophages in the lamina propria. However, PAS staining can give false-positive results because of other infections, for example with nontuberculous mycobacteria, and also false-negative results because of low bacterial load [9]. Therefore, every positive PAS result should be confirmed by an independent method. Here, molecular techniques such as PCR are, so far, irreplaceable for providing a direct, valid species diagnosis. Several in-house PCR protocols are now successfully used to detect T. whipplei DNA [10, 11]. In patients without gastrointestinal manifestation of classical Whipple’s disease, sample specimens from the clinically affected organs, e.g. heart valves, lymph nodes, synovial tissue, cerebrospinal fluid (CSF) or brain biopsies, may be PAS-positive, whereas duodenal biopsies remain negative [1]. PCR was suggested for screening stool and saliva samples as the prevalence and load of T. whipplei is far higher in Whipple´s disease patients than in healthy controls [4]. Here, however, positive PCR results are no proof of infection compared to the direct detection of T. whipplei DNA in affected organs. Analysis of peripheral blood is also possible, but a negative PCR result will not rule out infection [12]. As with all PCR assays, results need to be carefully interpreted as the assay is prone to laboratory contamination (especially nested PCR protocols) or false-positive results because of nonspecific reaction conditions or primer design. Importantly, some positive PCR results in the past have been shown to be due to cross-reactivity, e.g. with Actinomyces odontolyticus [13].
Improved diagnostics of T. whipplei
A break-through for the diagnosis of Whipple’s disease that is specific and less prone to contamination is modern real-time PCR [5, 14]. We evaluated a real-time PCR assay targeting T. whipplei-specific segments within the rpoB gene on test strains and over 1000 clinical specimens in a national reference laboratory [14]. This assay proved to be specific, sensitive and substantially faster than a conventional in-house assay. The protocol includes two specific hybridization probes and, to our knowledge for the first time in T. whipplei diagnostics, a melting curve analysis. Both are crucial for the robustness and reliability of the assay. This applies especially to polymicrobial samples, such as saliva or stool, which contain numerous uncultured bacterial species with unknown DNA sequences. Here, the problem of unexpected probe binding with false-positive results remains and, therefore, PCR results should always be interpreted in the context of clinical and histopathological findings. An initial diagnosis of Whipple´s disease should not rely on only one isolated PCR result, and a confirmatory PCR (using a different target sequence, sequence analysis of ribosomal RNA sequence or genotyping PCR) is mandatory. In inconclusive cases a second PCR with an independent sample specimen is recommended.
Emerging techniques for the detection of T. whipplei
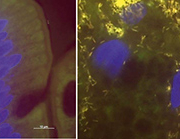
Besides PCR and PAS staining, additional methods such as immunohistochemistry or fluorescence in situ hybridization (FISH) are offered by specialized laboratories. These techniques are, as yet, not part of the routine work-up but provide promising insights. FISH uses fluorescently labelled probes that hybridize specifically with their target sequence in the intact bacterial cells (usually the 16S rRNA). Thus, FISH not only provides direct identification of T. whipplei but also visualizes the pathogen directly in the tissue context. Surprisingly, we found T. whipplei to be by far the most abundant cause of culture-negative endocarditis among the rare pathogens [15]. FISH revealed impressive infected areas in heart valves densely scattered with T. whipplei. In gut biopsies FISH reveals the amount and localization of single microorganisms in the tissue (Fig. 1). As with all microscopic techniques, however, FISH is less sensitive than PCR and will only give information on post-operatively obtained tissue and exclusively on the section investigated. Thus, a low bacterial load in the tissue might be missed. However, FISH is so far the only method bridging the gap between specific molecular biology and histopathology and, thus, might find broader application in the future.
Sampling for T. whipplei
Tissue specimens, such as small bowel biopsies in classical Whipple´s disease or samples of the affected organ in isolated T. whipplei infection, should be examined by PAS staining and PCR (Fig. 2). In the event of positive results, CSF should be tested by PCR to check for CNS involvement. For isolated T. whipplei infections gastrointestinal involvement should be controlled as well. Fluid samples, such as CSF, etc., should be examined by PCR.
For histological examination, PAS staining and FISH the samples should be fixed in 10% formalin and transported at room temperature. For PCR the samples need to be native (no formalin pre-treatment!) and can be transported at room temperature within one day. Samples can be stored for a few days at 4°C and should be kept at –80°C for long-term storage.
Conclusions
The recent development of real-time PCR protocols with hybridization probes for the specific detection of T. whipplei provides accurate and fast results in the challenging clinical situation of Whipple´s disease. However, due to the variety of clinical symptoms, asymptomatic carriage, isolated and systemic infection, as well as false positive and negative results all examinations need careful interpretation in specialized centres. Clinical and histopathological facts always have to be taken into account. Emerging techniques such as FISH might in the future close the gap between molecular biology and histopathology. Together clinical and microbiological expertise are the key to the fast and successful treatment of Whipple´s disease. Similarly, after initial diagnosis and initiation of treatment, it is highly recommended to follow each patient in specialized centres during and after antibiosis to keep relapses at bay.
Acknowledgements
We thank the Robert Koch Institute for its continuous support.
Funding Sources
This study was supported by the Robert Koch Institute (RKI). The epifluorescence microscope was a gift from the Sonnenfeld-Stiftung.
References
1. Moos V, Schneider T. Changing paradigms in Whipple’s disease and infection with Tropheryma whipplei. Eur J Clin Microbiol Infect Dis. 2011; 30: 1151–1158.
2. Raoult D, Birg ML, La Scola B, Fournier PE, Enea M, Lepidi H, et al. Cultivation of the bacillus of Whipple’s disease. N Engl J Med. 2000; 342: 620–625.
3. Lagier JC, Lepidi H, Raoult D, Fenollar F. Systemic Tropheryma whipplei: clinical presentation of 142 patients with infections diagnosed or confirmed in a reference center. Medicine 2010; 89: 337–345.
4. Fenollar F, Laouira S, Lepidi H, Rolain JM, Raoult D. Value of Tropheryma whipplei quantitative polymerase chain reaction assay for the diagnosis of Whipple disease: usefulness of saliva and stool specimens for first-line screening. Clin Infect Dis. 2008; 47: 659–667.
5. Fenollar F, Trani M, Davoust B, Salle B, Birg ML, Rolain JM et al. Prevalence of asymptomatic Tropheryma whipplei carriage among humans and nonhuman primates. J Infect Dis. 2008; 197: 880–887.
6. Martinetti M, Biagi F, Badulli C, Feurle GE, Müller C, Moos V et al. The HLA Alleles DRB1*13 and DQB1*06 Are Associated to Whipple’s Disease. Gastroenterology 2009; 136: 2289–2294.
7. Moos V, Schneider T. The role of T cells in the pathogenesis of classical Whipple’s disease. Expert Rev Anti Infect Ther. 2012; 10: 253–255.
8. Schneider T, Moos V, Loddenkemper C, Marth T, Fenollar F, Raoult D. Whipple’s disease: new aspects of pathogenesis and treatment. Lancet Infect Dis. 2008; 8: 179–190.
9. Müller SA, Vogt P, Altwegg M, Seebach JD. Deadly carousel or difficult interpretation of new diagnostic tools for Whipple’s disease: case report and review of the literature. Infection 2005; 33: 39–42.
10. Hinrikson HP, Dutly F, Nair S, Altwegg M. Detection of three different types of ‘Tropheryma whippelii’ directly from clinical specimens by sequencing, single-strand conformation polymorphism (SSCP) analysis and type-specific PCR of their 16S-23S ribosomal intergenic spacer region. Int J Syst Bacteriol. 1999; 49: 1701–1706.
11. Relman DA, Lepp PW, Sadler KN, Schmidt TM. Phylogenetic relationships among the agent of bacillary angiomatosis, Bartonella bacilliformis, and other alpha-proteobacteria. Mol Microbiol. 1992; 6: 1801–1807.
12. Marth T, Fredericks D, Strober W, Relman DA. Limited role for PCR-based diagnosis of Whipple’s disease from peripheral blood mononuclear cells. Lancet 1996; 348: 66–67.
13. Rolain JM, Fenollar F, Raoult D. False positive PCR detection of Tropheryma whipplei in the saliva of healthy people. BMC Microbiol. 2007; 7: 48.
14. Moter A, Schmiedel D, Petrich A, Wiessner A, Kikhney J, Schneider T et al. Validation of an rpoB gene PCR assay for detection of Tropheryma whipplei: 10 years’ experience in a National Reference Laboratory. J Clin Microbiol. 2013; 51: 3858–3861.
15. Geißdörfer W, Moos V, Moter A, Loddenkemper C, Jansen A, Tandler R et al. High frequency of Tropheryma whipplei in culture-negative endocarditis. J Clin Microbiol. 2012; 50: 216–222.
16. Mallmann C, Siemoneit S, Schmiedel D, Petrich A, Gescher DM, Halle E et al. Fluorescence in situ hybridization to improve the diagnosis of endocarditis: a pilot study. Clin Microbiol Infect. 2010; 16: 767–773.
The authors
Alexandra Wießner1, Annette Moter1* MD, Judith Kikhney1,2 PhD
1 Center for Biofilms and Infection, German Heart Institute Berlin, Berlin, Germany
2 Institut für Mikrobiologie und Hygiene, Charité University medicine Berlin, Berlin, Germany
*Corresponding author
E-mail: moter@dhzb.de