Quantitative proteomics: closing the gap between biomarker discovery and validation – the rate-limiting step
Many potential cancer biomarkers have been identified, however, the transfer of these biomarkers from discovery to clinical practice is still a process filled with pitfalls and limitations. To prove clinical utility and performance, these candidate biomarkers need to be reproducible, specific, sensitive, and validated using large sample cohorts, all of which require development of robust and highly sensitive multiplex protein assays.
by Dr G. M. Mustafa, Prof. J. R. Petersen, Prof. L. Denner Prof. F. A. Hussain and Prof. C. Elferink
Background
Although remarkable scientific and technological advances in medicine have been achieved, cancer incidences are still increasing worldwide with the ratio of deaths to new cancer cases remaining relatively unchanged at 49% [1]. The ability of physicians to effectively treat and cure cancer is directly dependent on their ability to detect cancer at an early stage. When cancer develops in an area of the body, such as an organ, as long as it hasn’t spread (metastasized), it may respond well to treatment and in some cases a cure may be possible. Many disease states, especially various types of cancer, can be better diagnosed by the aid of biomarkers, a key element of modern diagnostics and the value of which continuing to increase in modern medicine. As indicators of biological status, biomarkers, whether detected in blood, urine, or tissue, can be useful for the clinical management of various diseases. The changes in biomarker concentration have the potential to guide therapy in disease progression, prognosis and potentially be used to identify the stage of the cancer. Overall, protein biomarkers are needed to help our understanding of cancer biology and to improve our ability to diagnose, monitor and treat cancers.
Biomarker research
One of the goals of biomarker research is to discover and validate markers that can be used in clinical research applications such as patient stratification, diagnosis and therapeutic monitoring, or in pharmaceutical development to fully characterize the behaviour and efficacy of candidate drugs. The identification of prognostic, predictive or pharmacodynamic biomarkers can be carried out using genomic or proteomic technologies. Proteomic profiling of body fluids, such as serum, has potential as a sensitive diagnostic tool for early cancer detection. Serum provides a rich sample for diagnostic analyses because the expression and release of proteins (potential biomarkers) into the bloodstream occurs in response to specific physiological states. As human plasma has a 1010 dynamic range of proteins [2] with 22 proteins being responsible for 99% of the proteins identified, one of the challenges working with such a complex biological fluid is the difficulty in identifying medium and low abundance proteins. Thus, sample enrichment is a very important step in the discovery phase.
Biomarker validation – the rate-limiting step
Discovery platforms typically result in an extensive hit-list of candidate biomarkers. Important analytical and clinical hurdles must be overcome to allow the most promising protein biomarker candidates to advance into clinical validation studies. Over the last several years, many proteins have been proposed as potential biomarkers for various cancers without further evaluation of their clinical utility. The lack of follow-up is due, to a large extent, to the lack of an efficient technology for reproducible and accurate verification of these proteins as biomarkers in a specific disease state. Without proper validation, the identification of biomarkers is of minimal utility. To show clinical utility, biomarkers must be validated using a reliable assay with an independent cohort in a prospective or longitudinal study. A search of the scientific literature clearly indicates that most published biomarkers are inadequate for replacing an existing clinical test or that they are only useful for detecting disease in an advanced stage, where treatment success and/or survival rates are low. Traditionally, validation has been performed with immunoassays which require antibodies that are often unavailable or of poor quality. In addition the immunoassays may not adequately identify or differentiate between post-translational modifications. The process of generating antibodies is also time consuming and costly and is therefore not a practical solution for screening the hit-list of proteins derived from discovery.
Targeted mass spectrometry for multiplexed protein quantitation
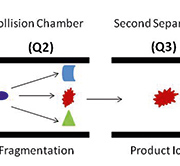
A targeted mass-spectrometry-based antibody-free platform for multiplexed protein quantitation fills the well-recognized gap between early biomarker discovery and final clinical assay. Stable-isotope-dilution multiple-reaction-monitoring mass spectrometry (SID-MRM-MS) is a novel technique whereby the quantification of a protein can be calculated from isotopically labelled peptide standards of known concentrations that correspond to the protein of interest [3]. With two to five unique peptides per protein, the concentration of the protein of interest can be accurately determined. Further, the technique can be multiplexed to allow for the simultaneous measurement of many proteins. We have validated this approach using serum samples to identify biomarkers in hepatocellular carcinoma and are confident that it also has the potential for biomarker discovery in other cancers [4]. This powerful workflow opens up new possibilities for biomarker research that may lead to faster, more robust, and improved clinical assays. Targeted proteomics workflows based on SRM and MRM on triple quadruple mass spectrometry platforms show the potential for rapid verification of biomarker candidates in plasma by using heavy isotope-labeled internal standards. This approach has the selectivity, reproducibility, and sensitivity for a range of multiplexed protein assays and has the potential for quantifying protein isoforms in addition to posttranslational modifications for which good quality antibodies often do not exist. The ability to rapidly quantify proteins in a highly multiplexed manner using MRM with internal standard peptides closes the gap between discovery and validation in the biomarker pipeline, which is the rate-limiting step in the biomarker world. Using a triple quadruple mass spectrometer, the first mass analyser (Q1) is set to only transmit the parent weight of a peptide from the target protein, the collision energy is then optimized to produce a diagnostic charged fragment of a peptide fragment in the second mass analyser (Q2), and the third mass analyser (Q3) is set to only transmit and identify this diagnostic peptide fragment (Fig. 1). The key strength of this work flow is that the protein assay development for hundreds of proteins can occur on the order of a month. Once identified, it just takes months to validate the low to sub-ng/ml concentration of candidate biomarkers in hundreds of blood samples. No target-specific reagents, such as antibodies, are needed and only a very small amount of sample (<500ng) is required for quantitation. In addition all potential biomarkers can be quantified in a single assay that is more accurate, rapid, and cost-effective. The capacity of SRM for multiplexed, high-throughput analysis, together with its sensitivity and quantitation, positions SRM as a promising application in medical screening [5, 6].
Summary
The type of clinical proteomics described here has important direct ‘bedside’ applications. We can foresee a future in which the physician will use these proteomic analyses at many points in the management of disease and drug discovery.
References
1. Yeom YI, Kim SY, Lee HG, et al. Cancer biomarkers in ’omics age. Biochip Journal 2008; 2(3): 160–174.
2. Anderson L. Candidate-based proteomics in the search for biomarkers of cardiovascular disease. J Physiol. 2005; 15: 23–60.
3. Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008; 4: 222.
4. Mustafa MG, Petersen JR, Ju H, Cicalese L, et al. Biomarker discovery for early detection of hepatocellular carcinoma in hepatitis C-infected patients. Mol Cell Proteomics 2013; 12(12): 3640–3652
5. Keshishian H, Addona TA, Burgess M, et al. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics (2007); 6(12): 2212–2229
6. Zhao Y, Jia W, Sun W, et al. Combination of improved 18O incorporation and multiple reaction monitoring: a universal strategy for absolute quantitative verification of serum candidate biomarkers of liver cancer. J Proteome Res. (2010); 9(6): 3319–3327
The authors
Gul M. Mustafa*1 PhD, John R. Petersen2 PhD, Larry Denner3 PhD, Feroze A. Hussain4 MD, and Cornelis Elferink1 PhD
1 Department of Pharmacology, University of Texas Medical Branch, Galveston, Texas, USA
2 Victory Lakes Clinical Laboratory, Department of Pathology, University of Texas Medical Branch, Galveston, Texas, USA
3 Department of Internal Medicine, University of Texas Medical Branch, Galveston, Texas, USA
4 Division of Gastroenterology and Hepatology, Department of Internal Medicine, University of Texas Medical Branch, Galveston, Texas, USA
*Corresponding author
E-mail: gmmustaf@utmb.edu