Artificial intelligence is the key driver for digital pathology adoption
by Dr Mustafa Yousif, Dr David S. McClintock and Dr Keluo Yao
Artificial intelligence (AI) has the potential to optimize anatomic pathology (AP) laboratory efficiency, enhance pathologists’ diagnostic skills, elevate case reimbursement and, ultimately, improve patient care. To realize this potential, AP laboratories must overcome the barriers to adopting digital pathology (DP) while DP vendors concurrently embrace whole slide imaging system standardization, interoperability and integration with AI platforms. We describe here how AI will drive DP adoption and overall add value to the practice of pathology.
Introduction to artificial intelligence and digital pathology
Artificial intelligence (AI) is defined as creating machines that think the way humans think, i.e. they can understand the world, make predictions, choose appropriate actions and perform judgmental processes associated with human intelligence [1]. Although this definition of strong/general AI lends itself well to science fiction, which often seems to progress to machines achieving intelligence far surpassing that of humans (super AI) and ultimately deciding to destroy the human race, the truth of AI’s current use in the world is fortunately much simpler. Instead of strong or super AI, the machine learning (ML) algorithms driving AI today are collectively known as weak, or narrow AI. They are focused algorithms designed to answer specific questions or solve distinct problems and are applied to narrowly defined subject domains, such as virtual assistants, predicting traffic patterns, self-driving cars, clinical decision support, etc.
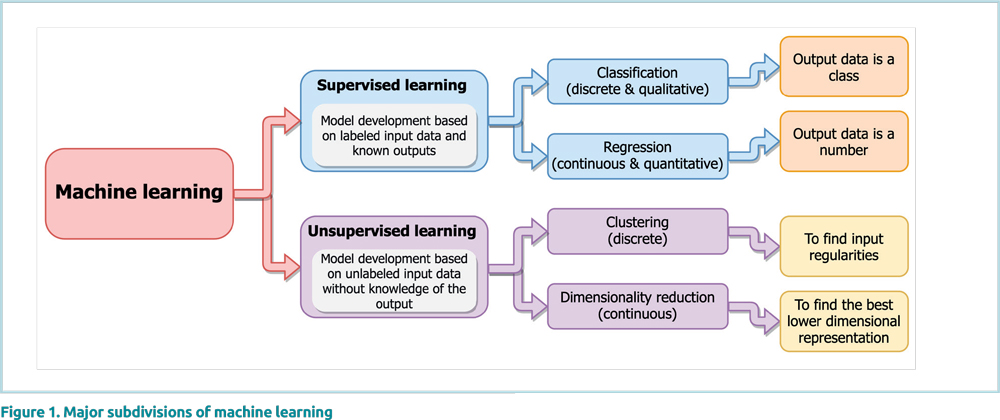
As mentioned above, ML algorithms form the underpinnings of modern AI – they minimize the manual input necessary to program an AI construct. In general, ML is a collection of different technological approaches allowing computers to solve problems without explicitly being programmed to learn and improve automatically through experience [2]. Although there are almost a hundred different ML methods available, the primary ML subdivisions most pertinent to pathology are shown in Figure 1 [3]. Of note, pathology is no stranger to AI, with many non-ML-based AI image analysis algorithms widely deployed in both anatomic and clinical pathology before the popularization of ML [4, 5]. However, it is only recently, with the advent of high-throughput digital pathology (DP), that AI has truly gained ground in anatomic pathology (AP) [6].
DP is best described as the tools and systems required to digitize pathology slides and other related pathology images, including not only the image data, but also all associated metadata, storage, analysis and enabling infrastructures [7]. Currently, DP is typically equated with whole slide imaging (WSI), with many groups using the terms interchangeably. Over the past 20 years, as technological and regulatory barriers have diminished, WSI has moved beyond its initial education and research use cases and has found significant clinical roles with primary diagnosis, secondary diagnosis (2nd opinions/consults), intraoperative consultation, multidisciplinary conferences, telepathology and telecytology.
DP advances and barriers
WSI serves as the foundational data platform, and is an absolute requirement for the application of AI in AP. Simply put, without digital pathology, there is no artificial intelligence in anatomic pathology (i.e. No DP = No AI). Fortunately, multiple recent advances in WSI have, in theory, paved the way for increased DP adoption by AP laboratories, such as (1) high-throughput scanning (e.g. output of 80–100 slides per hour per WSI device); (2) regulatory approvals for primary clinical use; (3) improved interoperability through imaging standards; and (4) increased pathologist acceptance of using virtual slides [8–10].
The reality surrounding DP adoption, however, is much more complex. WSI, in many ways, has a more complicated value proposition than other clinical imaging modalities (e.g. radiology) owing to its continued dependence on physical workflows (WSI still requires glass slides), substantial storage requirements (100s of TBs to PBs of annual data), diversity of primary and ancillary stains, use of multiple focal planes, reliance on pathologists as part of the clinical validation pathway, and a lack of non-pathology clinician awareness/acceptance. Further, even for simple implementations, the five-year total cost of ownership for DP can be estimated in the millions of dollars when all components of a WSI system are taken into account (WSI devices, image management system, storage, workstations and displays). Although some have shown positive return-on investment for DP implementation efforts [10–12], these studies for the most part are based on potential savings/projections and not on full accounting cost savings. Therefore, in order to see more widespread WSI adoption, a more tangible way of adding value through DP must be found.
AI is the key driver for DP adoption
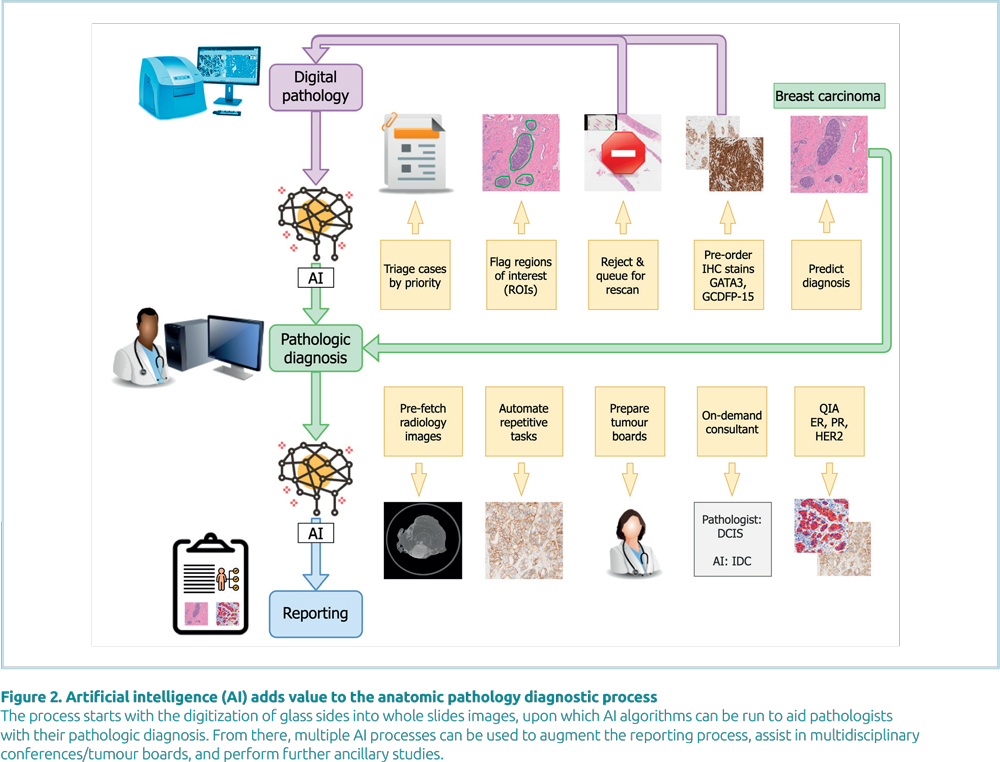
AI has the means to add significant value to AP by making the AP lab more efficient, automating repetitive tasks for pathologists, elevating case reimbursement and improving patient care. As Figure 2 demonstrates, there are many ways in which AI can be used at all stages of the pathology diagnostic process, including before, during and after the pathologist receives a patient’s whole slide images. Each of these optimizations can be correlated with time savings, cost savings, increased reimbursement and/or improved quality/patient safety (e.g. through measurable key performance indicators).
To realize this added value, however, it is not enough to simply scan one’s glass slides; instead, full interoperability and standardization of WSI is key. Currently, most WSI image management systems (IMS) in use clinically have the capability to interface with the AP laboratory information system in order to correctly identify patients/slides, collate virtual slides into cases and present the slides via an integrated viewer. Depending on the vendor, the IMS may also have the ability to integrate directly with specific AI platforms or to run specific AI algorithms.
Over the past 20 years, DP vendors have made great strides towards standardization and interoperability of WSI formats by conforming their devices and image outputs to the Digital Imaging and Communications in Medicine (DICOM) standard [13]. Although DICOM for WSI is still a work in progress, recent connectathons at national conferences have demonstrated the promise of true WSI interoperability [14]. Beyond DICOM, WSI format openness is also crucial for driving AP’s adoption of AI. Currently, the large number of competing WSI formats poses challenges for pathology AI algorithm development, with some ML techniques poorly generalizable across different image formats [15]. Fortunately, virtually all vendors have now made their image format technical documentation available publicly online.
AI algorithm development
Most AI systems have their roots in biomedical research using AP tissue, slides, and images. For example, in order to solve the problem of how to quickly evaluate large volumes of immunohistochemical (IHC) stained tissue with ER, PR, Her2, and Ki67, early research yielded high-throughput, semi-automated quantification algorithms for these specific biomarkers. Over time some of the platforms hosting these algorithms were commercial-ized, moving from offering single use case algorithms like quantification of IHC markers to sophisticated ML platforms with the ability for users to customize their use cases and develop, train and validate their own ML algorithms. Beyond the commercial systems, open source platforms have emerged with fully developed graphical user interfaces for developing custom image analysis algorithms using ML or non-ML methods. Finally, for algorithms of increasing complexity, development is probably better handled by Python or other similar programming languages with well-supported ML and image analysis software libraries, especially if the goals are future commercialization and integration into a WSI data pipeline.
Impact of AI on reimbursement and the practice of medicine
The impact AI will have on reimbursement for pathology services is also another significant consideration towards its future deployment. In the United States, the American Medical Association has officially recognized advances in AI and has updated the Current Procedural Terminology (CPT) codes to reflect these changes. For example, CPT codes 88365 to 88368 allow billing for computer-assisted in situ hybridization morphometric analysis while CPT code 88361 allows for an elevated charge for computer-assisted quantitative measurement of immunohistochemical markers. As a result of these special billing codes, dedicated FDA (U.S. Food and Drug Administration) approved/ cleared instruments with vendor validated algorithms are commercially available for breast biomarkers and Ki67 from several vendors. Further, the Center for Medicare and Medicaid Services (CMS) also provides alternative mechanisms for new, but costly, medical technologies. At the time of this writing, Viz ContaCT, a large vessel occlusion detection algorithm for computed tomography angiograms, received CMS New Technology Add-on Payment designation [16]. Although this specific algorithm does not pertain to pathology directly, it does represent a significant milestone in the quest for clinically-based AI algorithms becoming reimbursable in the USA, paving a similar pathway for AI algorithms in pathology.
Besides creating additional direct revenue generating charges, DP coupled with AI holds immense potential to revolutionize other areas of medicine, such as oncology and precision medicine. For example, pairing histopathology-based image analysis data with molecular/genomics data has the potential to yield new, unforeseen relationships when subjected to unsupervised ML techniques. This pairing has already started to pay off, with a study showing how mutation predictions for non-small cell lung cancer can be made using deep learning algorithms trained on histopathology images [17]. On a more practical level, simply automating and standardizing manual and subjective image analysis tasks may have downstream patient care effects besides increasing pathologist throughput or improving case turnaround times. Instead, automation and standardization of pathologist tasks such as the assessment of tumour necrosis, detection of lymphovascular invasion, quantification of mitotic figure counts per square area of tumour, or characterizing the full extent of fibrosis could affect clinically impactful measures like tumour grade, patient treatment options and prognosis.
Summary
DP has great potential for improving the practice of pathology, however there are real barriers limiting its adoption, the largest of which is typically cost. AI has the means to overcome these barriers by adding value to not only the DP process, but the entire pathology diagnostic process. By looking to implement both DP and AI together, one can more easily justify both of their adoptions and improve the practice of pathology.
The authors
Mustafa Yousif1 MD, David S. McClintock1 MD, Keluo Yao*2 MD
- Department of Pathology and Clinical Laboratories, University of Michigan, Ann Arbor, MI 48109, USA
- City of Hope National Medical Center, Department of Pathology, Duarte, CA 91010, USA
* Corresponding author
E-mail: kyao@coh.org
References
- Goodfellow I. Deep Learning Chapter 1 Introduction presented by Ian Goodfellow [Internet]. YouTube 2017 (https://www.youtube.com/watch?v=vi7lACKOUao&t=684s).
- Koza JR, Bennett FH, Andre D, Keane MA. Automated design of both the topology and sizing of analog electrical circuits using genetic programming. In: Gero JS, Sudweeks F (eds) Artificial intelligence in design ’96, pp151–170. Springer 1996. ISBN 978-94-010-6610-5.
- The world of machine learning algorithms – a summary infographic [Internet]. ERDataDoc blog 2016 (https://erdatadoc.com/2016/09/03/the-world-of-machine-learning-algorithms-a-summary-infographic/).
- Farris AB, Cohen C, Rogers TE, Smith GH. Whole slide imaging for analytical anatomic pathology and telepathology: practical applications today, promises, and perils. Arch Pathol Lab Med 2017; 141(4): 542–550.
- Yao K, Singh A, Sridhar K, et al. Artificial intelligence in pathology: a simple and practical guide. Adv Anat Pathol 2020; 27(6): 385–393.
- Cheng JY, Abel JT, Balis UGJ, et al. Challenges in the development, deployment & regulation of artificial intelligence (AI) in anatomical pathology. Am J Pathol 2020; S0002-9440(20)30508-3.
- Abels E, Pantanowitz L, Aeffner F, et al. Computational pathology definitions, best practices, and recommendations for regulatory guidance: a white paper from the Digital Pathology Association. J Pathol 2019; 249(3): 286–294.
- Zarella MD, Bowman D, Aeffner F, et al. A practical guide to whole slide imaging: a white paper from the digital pathology association. Arch Pathol Lab Med 2019; 143(2): 222–234.
- Hanna MG, Parwani A, Sirintrapun SJ. Whole slide imaging: technology and applications. Adv Anat Pathol 2020; 27(4): 251–259.
- Baidoshvili A, Bucur A, van Leeuwen J, et al. Evaluating the benefits of digital pathology implementation: time savings in laboratory logistics. Histopathology. 2018; 73(5): 784–94.
- Hanna MG, Reuter V, Samboy J, et al. Implementation of digital pathology offers clinical and operational increase in efficiency and cost savings. Arch Pathol Lab Med 2019; 143(12): 1545–1555.
- Ho J, Ahlers SM, Stratman C, et al. Can digital pathology result in cost savings? A financial projection for digital pathology implementation at a large integrated health care organization. J Pathol Inform 2014; 5(1): 33.
- Herrmann MD, Clunie DA, Fedorov A, et al. Implementing the DICOM standard for digital pathology. J Pathol Inform 2018; 9: 37.
- Clunie D, Hosseinzadeh D, Wintell M, et al. Digital imaging and communications in medicine whole slide imaging connectathon at digital pathology association pathology visions 2017. J Pathol Inform 2018; 9: 6.
- Zech JR, Badgeley MA, Liu M, et al. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS Med 2018; 15(11): e1002683.
- Hassan AE. New technology add-on payment (NTAP) for Viz LVO: a win for stroke care. J Neurointerv Surg 2020; doi: 10.1136/ neurintsurg-2020-016897.
- Coudray N, Ocampo PS, Sakellaropoulos T, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med 2018; 24(10): 1559–1567.