Bridging the omics gap: From clinical research to pathology
Matrix-assisted laser desorption/ionization (MALDI) imaging is a technique that is on the cusp of revolutionizing histopathology. This article discusses the benefits of MALDI imaging and how it is beginning to be used in oncology, with particular relevance to the challenging area of lipid analysis.
Background
Routine pathology requires standardized workflows and instruments that can identify markers of disease and inform therapeutic response while conserving limited sample material. New techniques often develop in clinical research applications ahead of them migrating into routine practice, such as where scientists are looking to understand the etiology of a disease or pharma development specialists are investigating biomarkers to stratify or follow patients in clinical trials.
Matrix-assisted laser desorption/ionization (MALDI) imaging mass spectrometry (MS) complements and expands the range of techniques available for providing vital spatial information about biomolecules in tissues. MALDI imaging requires only a single tissue section to map hundreds of biomolecules (proteins, lipids and glycans, for example) in a label-free, untargeted manner.
This article highlights the value of MALDI imaging as it is established in clinical research and discusses potential applications in pathology. We consider, in particular, its impact in cancer research and disease management, and introduce recent advances in technology, including trapped ion mobility spectrometry (TIMS) and laser-induced postionization (PI), that broaden the depth of molecular information available and look set to drive the transition into patient-care benefits.
Molecules in spatial context
Over several decades, traditional diagnostic histopathology of tissue sections has been gradually augmented with the use of molecular labels and probes as specific molecular markers of disease are discovered. Converging with the knowledge provided by the Human Genome Project in the 1990s and early 2000s, The Cancer Genome Atlas (TCGA) programme that began in 2006, and the development of accessible sequencing technology, reverse transcription (RT-)PCR, immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH) methodologies, a new discipline – clinical molecular diagnostics – was born.
Looking back on these developments, one 2017 review concluded that “Pathologists will become pilots for precision cancer therapy through their unique ability to combine morphological and molecular findings” [1].
The goal then, as now, was to connect morphological information with genetic and molecular insights. For example, early work in cancer demonstrated the efficacy of tyrosine kinase inhibitors erlotinib and gefitinib in the treatment of EGFR-mutated lung adenocarcinomas and forced pathology departments to adopt EGFR mutation testing. This helped to establish the paradigm that led, ultimately, to today’s emphasis on omics and the development of spatial omics techniques.
Current tools – and ongoing challenges
Now, with spatial information seen as a vital component when analysing the microenvironment of disease in tissues and determining the location and interactions of cellular components that dictate disease outcome, a suite of diagnostic tools has become established that includes IHC, spatial transcriptomics, and imaging mass cytometry.
While the use of these techniques ranges over many disease pathologies – in neurology and cardiology, for example – oncology has seen the widest and deepest application to date. All bring useful targeted information about proteins in tissues; however, each has its own challenges (Fig. 1). An important general limitation for all three tools is an inability to capture the variety of post-translational modifications (PTMs) in the proteome, or visualize the lipidome and metabolome, all of which would offer a broader molecular insight and stronger basis for classification compared to considering just the pre-translational proteome.
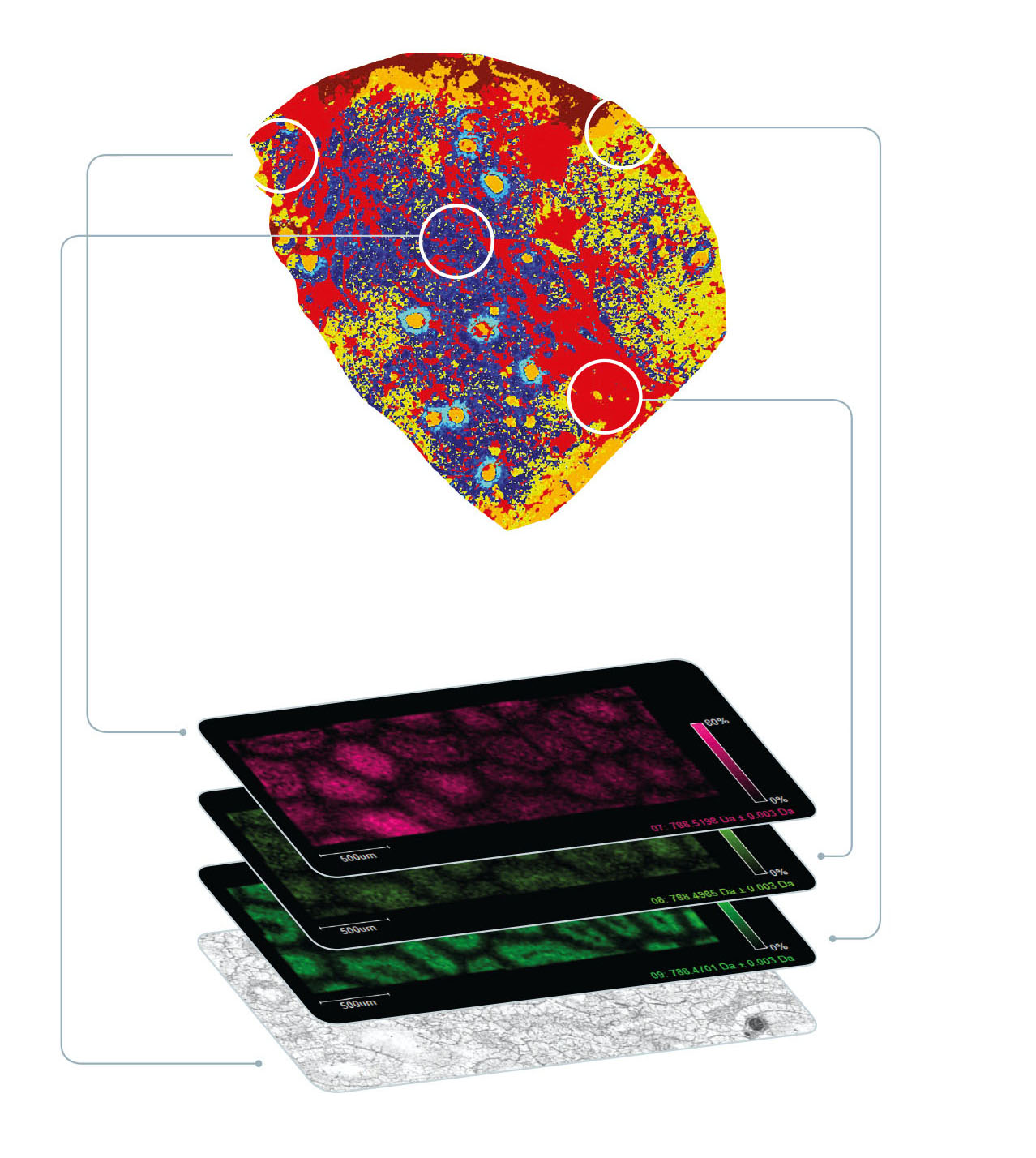
In contrast, MALDI imaging offers a label-free tool that captures information about the spatial proteome but additional spatial omic signatures that are unique to the local cell neighborhood. No prior knowledge of the compounds is required – the technique provides true untargeted molecular analysis in spatial context. Importantly, tumour-associated biomolecules that are missed at the gene level can be visualized. From a practical point of view, the MALDI imaging workflow is compatible with standard histological procedures, it maintains spatial resolution at around 10 μm, and the tissue section under test is preserved for further study.
Many researchers agree that moving beyond protein biomarkers is important for next-stage clinical understanding, with one recent publication noting, for example, that “Lipids play a significant role in the manifestation of cancer. However, research into lipid biomarkers of cancer is still in its infancy” [2].
MALDI imaging offers a powerful complementary technique to discover and spatially map such important features at a deeper molecular level (Fig. 2).
Pushing into routine oncology
The advantages of MALDI imaging have significant diagnostic potential. In-depth spatial proteomic, lipidomic, and metabolomic insights that complement traditional genomic and transcriptomic methods can, for example, help identify new predictive or prognostic biomarkers, and classify heterogeneous tumour subpopulations to give important contextual clues to tissue-level communication networks that are integral to cancer growth and treatment success.
Many recent studies demonstrate the advantage of integrating MALDI imaging with traditional techniques for tissue pathology applications. For example, Yagnik et al. reported the development of a new method based on novel photocleavable mass-tags (PC-MTs) for facile antibody labelling, which enables highly multiplexed IHC based on MALDI mass spectrometric imaging (MALDI-IHC [3]). Their conclusions were that the new combination shows promise for use in the fields of tissue pathology, tissue diagnostics, therapeutics, and precision medicine [3].
Work published in 2019 by Randall et al. demonstrated how MALDI imaging of lipids and metabolites in tissue samples accurately reflected a patient’s prostate cancer stage, as defined by traditional histologic evaluation using the Gleason score. This is the current standard of care; however, it is a time consuming process that is prone to intra-/inter-observer variability, and provides no information about altered metabolic pathways or altered tissue architecture. They concluded that MALDI imaging could be used as a potential clinical tool to support more objective and faster diagnosis [4].
To assess the suitability of MALDI imaging as a front-line technique, Basu et al. recently reported on ‘real-time’ assessment of tumour margins. Their goal was to discriminate surgical resection specimens from patients in a workflow that was rapid enough to be applied in clinical pathology [5]. By adapting various stages of a conventional 30-minute protocol for MALDI imaging, they were able to develop a reliable and reproducible 5-minute workflow that they concluded placed MALDI imaging firmly in the realm of routine clinical decision-making. See Fig. 1 in Basu et al. for their comparison of conventional workflows and rapid MALDI imaging (https://doi. org/10.1038/s41698-019-0089-y) [5]. Furthermore, they suggest that, by using an artificial intelligence (AI) step in the workflow, the MALDI imaging data could be analysed directly, without visual review, using previously established machine trained models.
Looking ahead
Technology developments are continuing, and two recent breakthroughs are now also being applied to MALDI imaging. First, ion mobility separation (IMS) has greatly broadened the range of biomolecules that can be analysed by pre-separation ahead of mass analysis. Of the many IMS technologies that exist, TIMS offers a number of benefits for MALDI imaging as well as traditional omics, which was demonstrated in a joint project between Bruker and the University of Maastricht, The Netherlands, that illustrated how MALDI-guided spatial omics uncovers proteomic diversity in lipid-segmented subpopulations of breast cancer [6].
Second, novel laser-induced PI technology has delivered a quantum leap in MALDI imaging sensitivity, by up to three orders of magnitude. This has now been applied to a dual MALDI-electrospray ionization (ESI) mass spectrometer, combining high mass accuracy with advanced MALDI imaging for the separation and identification of analytes such as lipids and glycans in complex mixtures [7]. Currently, this exciting development is being explored in research projects, but it is easy to see how this could, in time, offer next-level performance to pathology applications too.
Conclusion
Clinical research has led the way in utilizing MALDI imaging technology, capitalizing on the powerful label-free analytical tool that can fill in the broad gaps that spatial transcriptomics and genomics leaves in molecular investigations on tissue samples. It can provide valuable information about protein modifications after gene expression and visualizes additional compounds such as, for example, metabolites, glycans and lipids that all play a role in disease pathology.
Crucially, it maintains the spatial relationships of analytes in tissues, allowing improved translational and clinical insights. Recent literature provides a compelling body of evidence for the consideration of MALDI imaging in routine pathology applications, and many see the potential of this technology to provide a top-down, disease-centric pathological view of tissues that can inform therapeutic strategies, support diagnosis and improve patient outcomes.
Further developments to the technology will provide faster measurement speeds, increased sensitivity without compromising spatial resolution, and even deeper molecular content – important factors that look set to accelerate the adoption of MALDI imaging in the routine clinical environment.
The author
Shannon Cornett PhD, Mass Spectrometry
Applications Development Manager, Bruker
Daltonics Bruker Corporation, MA 01821, USA
Email: Marketing.bdal.bre@bruker.com
For further information visit Bruker MALDI
Imaging (https://www.bruker.com/en/applications/academia-life-science/imaging/maldi-imaging.html)
References
1. Müllauer L. Milestones in pathology-from histology to molecular biology. Memo 2017; 10(1): 42–45 (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5357255/)
2. Stromberg LR, Lilley LM, Mukundan H. Advances in lipidomics for cancer biomarker discovery. In: Issaq HJ, Veenstra TD (eds) Proteomic and
metabolomic approaches to biomarker discovery, 2nd edn. Elsevier 2020; pp421–436. ISBN: 978-0128186077.
3. Yagnik G, Liu Z, Rothschild KJ, Lim MJ. Highly multiplexed immunohistochemical MALDI-MS imaging of biomarkers in tissues. J Am Soc Mass
Spectrom 2021; 32(4): 977–988 (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8033562/).
4. Randall EC, Zadra G, Chetta P, Lopez BGC, Syamala S, et al. Molecular characterization of prostate cancer with associated Gleason score using mass
spectrometry imaging. Mol Cancer Res 2019; 17(5): 1155–1165 (https://mcr.aacrjournals.org/content/17/5/1155.long).
5. Basu SS, Regan MS, Randall EC, Abdelmoula WM, Clark AR, et al. Rapid MALDI mass spectrometry imaging for surgical pathology.
NPJ Precis Oncol 2019; 3: 17 (https://doi.org/10.1038/s41698-019-0089-y).
6. Oetjen J, Hebeler R, Dewez C, Henkel C, Balluff B, et al. LCMS 166: MALDI guided SpatialOMx uncovers proteomic profiles in tumor subpopulations of breast
cancer. Bruker Daltonics 2020 (Application Note; https://www.bruker.com/en/applications/academia-life-science/imaging/maldi-imaging/SpatialOMx.html).
7. Soltwisch J, Heijs B, Koch A, Vens-Cappell S, Höhndorf J, Dreisewerd K. MALDI-2 on a trapped ion mobility quadrupole time-of-flight instrument for
rapid mass spectrometry imaging and ion mobility separation of complex lipid profiles. Anal Chem 2020; 92(13): 8697–8703.