Clinical application of NGS – ensuring quality
Advances in Next Generation Sequencing (NGS) are bringing much higher throughput and rapidly reducing costs, whilst facilitating new mechanisms for disease prediction. Consequently, the clinical applications of NGS technologies are continuing to develop, with the potential to change the face of genetic medicine [1].
by Hannah Murfet (BSc, PCQI), Product Quality Manager, Horizon Discovery
Applications of NGS in a clinical context are varied, and may include interrogation of known disease-related genes as part of targeted gene panels, exome sequencing, or genome sequencing of both coding and non-coding regions. However, as NGS moves further into the clinic, care must be taken to ensure high levels of quality assurance, rigorous validation, recording of data, quality control, and reporting are maintained. [1] [2]
Guidelines specific to NGS are beginning to emerge and to be adopted by clinical laboratories working with these technologies, in addition to those mandated by clinical accreditation and certification programmes. In this article we give an overview of the specific guidance set out by the American College of Medical Genetics and Genomics in its September 2013 report ‘ACMG clinical laboratory standards for next-generation sequencing’, and the New York State Department of Health’s January 2014 document ‘Next Generation Sequencing (NGS) guidelines for somatic genetic variant detection’.
Quality Assurance
Quality assurance (QA) in the clinical context comprises maintenance of a desired level of quality for laboratory services. Typically, quality management systems take a three tier hierarchy. At the highest level the policies define the organisation’s strategy and focus. Underneath this sit the procedures, which define and document instructions for performing business/quality management or technical activities. Underpinning both of these tiers are accurate records.
In the case of New York State Department of Health guidelines, there is clear focus on the requirement for SOPs, which can be broken down into two levels. The first level states the required flow of information, demonstrating the sequence of events, and associated responsibilities or authorities. The first level procedures are best kept at a relatively high level, and may reference more specific and detailed level two processes.
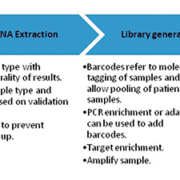
Testing sequences may be incorporated into one or more level one processes, depending on the complexity of the clinical laboratory’s operations. An overview of the typical testing sequence is shown in the figure below.
Level two processes are best documented as clear ‘how to’ guides, detailing all responsibilities, materials and procedures necessary to complete the activity. For laboratory-focused activities, validation study inputs and outputs can establish clear and consistent protocols, supporting training and laboratory operation.
Accurate record keeping should include which instruments were used in each test, as well as documentation of all reagent lot numbers. Any deviations from standard procedures should be recorded, including any corrective measures [1]. Templates may be generated to ensure consistency in output records for both testing and reporting.
In addition to documented processes, implementation of predetermined checkpoints or key performance indicators should be included to permit the monitoring of QA over time. Once established, these may act as a trigger for assay drift, operator variability, or equipment issues.
In the US, compliance to the HIPAA Act (Health Insurance Portability and Accountability Act) must be implemented to ensure traceability and protection of patient data, and many authorities mandate record retention periods, including CLIA who dictate that records and test reports must be stored for at least two years [1].
Clinical laboratories may look to further certification to ensure tight QA, such as the implementation of ISO 15189, especially in countries where no formal accreditation schemes are in place. [3]
Validation
Validation involves the in-depth assessment of protocols, tests, materials and platforms, providing confidence that critical requirements are being met. Test development and platform optimization should include factors such as determination of sample pooling parameters, and use of synthetic variants to create a strong data set, to compare tools and optimize the workflow. Validation of each entire test should be undertaken, using set conditions for sensitivity, specificity, robustness and reproducibility. It should be noted that the first test developed may naturally carry a higher validation burden than subsequent tests developed for the same platform. Platform validation and quality management are also vital. [1,2]
Specific validation requirements for NGS as set out by the New York State Department of Health are listed below. These guidelines may be used as a basic checklist for coverage, or to supplement more general accreditation or certification requirements, e.g. those required by CLIA or ISO 15189. [1]
- Each reportable variant does not require confirmation every time it is encountered, as long as the variant and target area (gene) containing it was rigorously validated
- Accuracy and validity of the bioinformatics must be demonstrated
- Anything that is not exclusively based on a FDA-approved assay is considered to be a laboratory developed test and will require full validation over verification
- Commercially available materials must be validated by the laboratory for use as a diagnostic tool where there are no clinical indications for use
- Validation of a single version of all analyses software
- Performance characteristics for each sample type must be established (e.g. FFPE)
- Performance characteristics for each type of variant in the assay must be established, and each type of detection should be validated separately (e.g. SNV or structural variants)
Data
NGS has the potential to create huge amounts of data, meaning that accurate and efficient systems for data storage and collection are more essential than ever. Data protocols are generally established through the validation stages, then monitored at predetermined checkpoints with key performance indicators to ensure consistency and accuracy of service provision.
The list below gives an overview of NGS specific data requirements from the New York State Department of Health. [1]
Accuracy
- Validation, including minimum 50 patient samples with representation for material type (e.g. FFPE), and variants across target areas, confirmed by an independent reference method
- Minimum 10 positive samples for each type of variant
- Recommended approach – sequence a well characterised reference sample to determine specificity
- If vigorous validation of reported variants has not been completed in the original studies, ongoing confirmation by independent reference methods must be performed until at least 10 reference points have been independently validated
- A disclaimer must be used where incidental findings of unknown significance are included, where there is no established confirmatory assay. The disclaimer must clearly state that the variant has not been verified
Robustness
- Robustness is the likelihood of assay success. Adequate quality control measures must be in place to determine success of techniques such as extraction, library preparation or sequencing
Precision
- Precision is related to within-run control
- For each type of variant a minimum of 3 positive samples containing variants near the stated sensitivity of the assay must be analysed in triplicate in the same run using different barcodes
- Renewable reference samples can be used to determine the analytical validity of the test. These can establish baseline data to which future modifications can be compared
Repeatability and Reproducibility
- Repeatability and reproducibility is related to between-run controls, to determine ability to return identical results under identical (repeatability) or changed (reproducibility) conditions
- For each type of variant a minimum of 3 positive samples containing variants near the stated sensitivity of the assay must be analysed in three separate runs, using different barcodes on different days, by two different technologists where possible
- If multiplexing samples with distinct barcodes, it must be verified that there is no cross talk and that all target areas and variants are reproducible, independent of which patient/barcode combination is used
- It is useful to consider instrument-instrument variability as well as inter-operator variability. Parameters for expected reproducibility should be established, and would typically be around 95-98%
Analytical Sensitivity and Specificity
- Sensitivity and specificity refer to positive and negative percentage variability respectively, when compared to gold standard
- All types of variants in three target areas with consistently poor coverage should be interrogated, as well as three target areas with consistently good coverage. These can be established with defined mixtures of cell line DNA (not plasmids), but must be verified with 3 – 5 patient samples
- The limit of detection should be established
- Confidence intervals for variant types must be determined
A minimum data set is expected, to establish key performance characteristics, including: base calling; read alignment; variant calling; and variant annotation.
Quality Control
In contrast to quality assurance where the infrastructure for quality is established to maintain the right service, quality control addresses testing and sampling to confirm outputs against requirements. Quality control takes place across all aspects of a process from reagents used, to software and in-assay controls.
Quality control of reagent lots is best implemented at the point of goods inspection. A clear label should be placed on the reagent under inspection, and testing performed to validate/confirm analytical sensitivity. Quality control of software updates can be handled through a version control and impact assessment process. All re-validation must be clearly documented and demonstrate consistency in analytical sensitivity.
Sample identity confirmation is essential, especially if samples are pooled. Proficiency testing protocols must be established to allow for execution as required by clinical accreditation bodies (such as CLIA). Quality control stops may be added to laboratory process before the sequencing run, to the run itself and at the end before data analysis.
Use of control materials /reagents at all stages of the sequencing procedure supports quality control. No Template Controls (NTC) should be used at all amplification steps; a negative Control should be used upon initial validation, and periodically thereafter; and a Positive / Sensitivity Control should be used in each sequencing run. [1]
Several different QC protocols may need to be followed, and quality control measures applied can vary depending on chosen methods and instrumentation, but they should always include procedures to identify sample preparation failures and failed sequencing runs. Documentation for QC protocols is best detailed in the relevant SOP.
Reports
Specific requirements around the generation, approval, issue and re-issue of reports are included as part of accreditation programmes, such as CLIA, and standards certifications, such as ISO 15189. The most essential reporting requirements related to NGS are as follows [1,2]:
- The laboratory director is responsible for designing advantages and limitations of test offerings, ensuring healthcare providers can make informed decisions
- Turnaround times for reports should ensure there are clear requirements for NGS test prioritisation, and should be clinically appropriate
- All detected somatic variants should be recorded in a report, identifying each variant’s significance
- Incidental findings including clinical relevance should be recorded
- Limitations of the assay should be identified and reported on, including for which target areas the assay lacked sufficient coverage to confidently determine mutational status
- Information comparing the level of exome vs. genome sequencing to an available disease specific panel test should be included
Conclusions
While complete understanding of the clinical implications of some variants is still to be fully understood, there are clear prospects emerging for NGS to support further development and adoption of companion diagnostics. As the overall picture for NGS evolves, sell-defined guidelines are being developed for everything from quality assurance to reporting. It is expected that guidance and certification will continue to develop as NGS becomes an ever more common technology within the clinical laboratory.
References
1. American College of Medical Genetics and Genomics. (2013, September). ACMG clinical laboratory standards for next-generation sequencing.
2. New York State Department of Health. (2014, January). “Next Generation” Sequencing (NGS) guidelines for somatic genetic variant detection.
3. Horizon Discovery. (n.d.). ISO 15189: A Standard of Yin and Yang.