Detection of circulating tumour cells from peripheral blood of breast cancer patients via real-time PCR
As the appearance of circulating tumour cells in the peripheral blood of breast cancer patients is linked to a worse prognosis for overall survival and treatment efficiency, their detection and characterization will have a high impact on cancer therapy, opening roads to a more personalized treatment.
by Dr U. Andergassen, Dr A. C. Kölbl, Prof. K. Friese and Prof. U. Jeschke
Circulating tumour cells
Already in 1869 the occurrence of cancer cells in the peripheral blood of a metastatic cancer patient was described by Thomas Ashworth. Nowadays it is well known, that cells dissolve from primary epithelial tumours such as breast, lung, colon or prostate cancer, enter circulation and travel via the blood stream or lymphatic system throughout the whole body. If these cells [termed circulating tumour cells (CTCs)] leave circulation, they can settle at other sites in the body and are then considered to be the main reason for the generation of remote metastasis. Their appearance is linked to a poorer outcome of cancer therapy and to a worse prognosis for overall survival. Therefore, the detection of CTCs in peripheral blood [and of disseminated tumour cells (DTCs) in bone marrow] was already included into the international tumour staging systems.
Unfortunately the detection of CTCs is still a technical challenge, as the number of tumour cells in the blood stream is rather small (1 in 106–7 blood cells). To date, there is only one FDA-approved system for CTC detection, at least in the metastatic situation. This is the Cell Search® system (Veridex LLC.), which is based on immunomagnetic enrichment and simultaneous staining of tumour cells of epithelial surface markers, the cytokeratins. The huge disadvantage of this system is that it is rather expensive and, therefore, not yet routinely used in the clinic.
Real-time PCR in cancer cell detection
Another promising approach for CTC detection could be a real-time PCR-based method. The principle of this methodology is that breast-cancer CTCs are derived from an epithelial tumour, and, therefore, express a panel of epithelial cell genes. The surrounding blood cells in contrast are of mesenchymal origin, showing different gene expression profiles. Thus, it can be assumed that tumour cells are present in a given blood sample if the expression of epithelial genes is higher than in a negative control sample.
Real-time PCR measures gene expression levels by detecting an increase of fluorescence due to the incorporation of fluorescent reporter molecules into the newly synthesized DNA molecules during the PCR reaction. If a gene is highly expressed, a lot of mRNA of this gene is present, meaning plenty target for PCR reaction is available and thus influencing the fluorescence level measured at the end of each amplification cycle. The time point when fluorescence reaches a certain threshold is called the Ct-value, and this is the basis of the calculation of relative gene expression values by the 2-∆∆Ct-method [1]. In brief: the average Ct-value of a gene of interest is related to the average Ct-value of a reference gene. The resulting value is called the ΔCt-value. In the next step, this ΔCt-value is set in reference to the ΔCt-value of the same gene in the reference sample, rendering the so called ΔΔCt-value. The formula 2–ΔΔCt is then used to calculate relative quantification (RQ) values. RQ values >1 show an upregulation of the gene of interest, values <1 mean that the gene is downregulated.
Spiking experiments
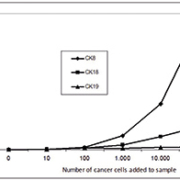
The first step towards a real time PCR based quantitative cancer diagnosis is to create calibration curves for the used marker genes to evaluate the number of cancer cells exhibited at a certain level of gene expression in a blood sample. Therefore, blood samples of healthy donors, to which a certain number of cells from a breast-cancer cell line were added, were used to create standard curves. For this evaluation different breast-cancer cell lines were used (Cama-1, MCF-7, MDA-MB231 and ZR-75-1), and real-time PCR was carried out for Cytokeratin 8, 18 and 19 as marker genes [2, 3]. Cancer cells were added in rising numbers and calibration curves could be drawn [Fig. 1], showing an increase in gene expression level from 10 cells added to a blood sample upwards, meaning that even a small number of cancer cells in the blood (resembling the ‘real’ conditions, with 1 CTC per106–7 surrounding blood cells) can be detected by this methodology.
PCR marker genes for CTC detection
As CTCs in the blood are rare, PCR marker genes have to be selected as accurately as possible. The first choice are the Cytokeratin (CK) genes 8, 18 and 19, as they are also used in the routinely applicated APAAP-staining, which is a histochemical detection method for CTCs. The cytokeratin family members are characteristic epithelial cell markers and only weakly expressed in blood cells, rendering them potentially useful for PCR-based detection of CTCs.
Three other genes (BCSP, MGL, Her2) were selected and used in an approach to detect differences in gene expression between normal individuals and adjuvant and metastatic breast-cancer patients [4]. Mammaglobin (MGL) is a gene which is only expressed in the adult mammary gland and is known to be upregulated in breast cancer [5]. Breast cancer specific protein (BCSP) is highly expressed in advanced infiltrating breast cancer and is a marker for recurrence of the disease and formation of metastases [6], and c-erbB2 (Her2) was used, because it is over-expressed in 20% of breast cancers and is also responsible for the aggressiveness of the tumour [7].
These markers were comparatively analysed in blood samples withdrawn from adjuvant and metastatic breast-cancer patients during surgery. The gene expression levels of adjuvant as well as metastatic breast-cancer patients were normalized to levels in blood samples from 20 healthy donors, considered as a negative control group. Differences in gene expression between the three sample groups were detected [Fig. 2] and it was attempted to find a signature of marker genes for CTCs in breast cancer by real-time PCR.
From the experiments, it could be concluded that cytokeratin genes seem to be the most promising markers for the detection of CTCs from peripheral blood of breast-cancer patients with reverse-transcription real-time PCR. The most suitable marker of the cytokeratin array is apparently CK8, rendering most expression values >1.
MGL, BCSP, and Her2 mRNA show few expression values >1 as well in adjuvant as in metastatic patients. Altogether, higher amplitudes for these three genes were detected in the adjuvant setting. CTCs can be detected from peripheral blood by real-time-PCR, using the cytokeratin markers, especially cytokeratin 8.
In contrast to these findings are the results published by Obermayr et al. 2010 [8], who found an overexpression of MGL/hMAM in 39% of the examined advanced breast cancer cases. But they also conclude that using more marker genes for CTC detection results in a higher percentage of detected cancer cases. The same findings were obtained by [9], who also used a real-time PCR-based approach for CTC detection. They used CK19, SCGB2A2, MUC1, EPCAM, BIRC5 and Her-2 as marker genes and found a high sensitivity and specificity (56.3% and 100% respectively).
Additionally CK20 was identified as a promising marker gene [10] and seems to be correlated with the aggressiveness of the tumour. To further improve the detection of CTCs by real-time-PCR, more marker genes need to be tested; promising candidates are, for example, MMP13 [11], UBE2Q2 [12],
Nectin-4 [13], and ALDH [14].
Future directions for cancer therapy
Real-time PCR-based techniques were already used for solid tumour profiling and are considered to be objective, robust and cost-effective molecular techniques that could be used in routine cancer diagnosis. In future, a real-time PCR assay for the detection of circulating tumour cells from peripheral blood could find its way into modern medicine. This would be advantageous for the patient by limiting the number of invasive procedures, such as biopsies or bone marrow aspirations, that have to be undertaken to produce samples for analysis.
Furthermore by implication of more marker genes a characterization of tumour cells could be pursued, which already gives hints towards a cancer prognosis, as for example Bölke et al. described, that the expression of certain genes is correlated to advanced breast cancer stages [15]. A better knowledge of cancer properties in turn will help to apply a more personalized therapy, side effects can be reduced and treatment efficiency will strongly increase.
References
1. Livak KJ, Schmittgen TD. Methods 2001; 25(4): 402–408.
2. Zebisch M, Kolbl AC, Schindlbeck C, Neugebauer J, Heublein S, Ilmer M, Rack B, Friese K, Jeschke U, Andergassen U. Anticancer Res 2012; 32(12): 5387–5391.
3. Zebisch M, Kölbl AC, Andergassen U, Hutter S, Neugebauer J, Engelstädter V, Günthner-Biller M, Jeschke U, Friese K. Biomedical reports; accepted for publication 2012.
4. Andergassen U, Hofmann S, Kolbl AC, Schindlbeck C, Neugebauer J, Hutter S, Engelstadter V, Ilmer M, Friese K, Jeschke U. Int J Mol Sci 2013; 14(1): 1093–1104.
5. Fleming TP, Watson MA. Ann N Y Acad Sci 2000; 923: 78–89.
6. Wu K, Weng Z, Tao Q, Lin G, et al. Cancer Epidemiol Biomarkers Prev 2003; 12(9): 920–925.
7. Kim YS, Konoplev SN, Montemurro F, Hoy E, Smith TL, et al. Clin Cancer Res 2001; 7(12):4008–4012.
8. Obermayr E, Sanchez-Cabo F, Tea MK, Singer CF, Krainer M, et al. BMC Cancer 2010; 10: 666.
9. de Albuquerque A, Kaul S, Breier G, Krabisch P, Fersis N. Breast Care (Basel) 2012; 7(1): 7–12.
10. Tunca B, Egeli U, Cecener G, Tezcan G, Gokgoz S, Tasdelen I, et al. Tumori 2012; 98(2): 243–251.
11. Chang HJ, Yang MJ, Yang YH, Hou MF, Hsueh EJ, Lin SR. Oncol Rep 2009; 22(5): 1119–1127.
12. Nikseresht M, Seghatoleslam A, Monabati A, et al. Cancer Genet Cytogenet 2010; 197(2): 101–106.
13. Fabre-Lafay S, Garrido-Urbani S, Reymond N, et al. J Biol Chem 2005; 280(20): 19543–19550.
14. Dontu G. Breast Cancer Res 2008; 10(5): 110.
15. Bolke E, Orth K, Gerber PA, Lammering G, Mota R, et al. Eur J Med Res 2009; 14(8): 359–363.
The authors
Ulrich Andergassen* MD, Alexandra C. Kölbl PhD, Klaus Friese MD, and Udo Jeschke PhD
Klinik und Poliklinik für Frauenheilkunde und Geburtshilfe, Ludwig Maximilian University of Munich, Munich, Germany
*Corresponding author
ulrich.andergassen@med.uni-muenchen.de