Diagnosis of SARS-associated coronavirus
Coronaviruses are a group of positive sense, single-stranded RNA viruses that infect humans and animals. In a short period of time the SARS-associated coronavirus was identified and initial laboratory protocols for diagnosis of SARS were disseminated. The need for the early diagnosis of SARS is vital due to the difficulty in clinically diagnosing this infection and its rapid nosocomial transmission.
by Dr Hoon H. Sunwoo and Dr Arivazhagan Palaniyappan
Clinical background
Severe acute respiratory syndrome (SARS) is a life-threatening viral respiratory illness caused by a coronavirus known as SARS-associated coronavirus (SARS-CoV, but usually shortened to SARS). The SARS-CoV is associated with a flu-like syndrome, which may progress into pneumonia, respiratory failure, and sometimes death. It is believed that SARS-CoV originated in the Guangdong Province in southern China and the virus has subsequently spread around the world. China and its surrounding countries have witnessed the greatest numbers of SARS-related cases and death.
SARS history is short. SARS-CoV was first reported in 2002 in Asia and cases were reported until mid-year 2003. According to the World Health Organization (WHO), as of July 2003, a total of 8437 people worldwide became ill and 813 died during the SARS outbreak or epidemic. Illness was reported in more than 30 countries and on 5 continents. This new emerging disease represented the most recent threat to human health as it has been reported to be highly contagious. Infection with the SARS-CoV causes acute respiratory distress (severe breathing difficulty) and sometimes death.
SARS-CoV Diagnosis
Three major diagnosis methods are currently developed (i) viral RNA detection using quantitative reverse transcription (RT)-PCR, (ii) antibody detection using indirect fluorescence assay (IFA), and (iii) using both recombinant nucleocapsid protein (NP) and culture extract of SARS-CoV–based enzyme-linked immunosorbent assay (ELISA). ELISA based antibody detection tests with recombinant antigens are well known to offer higher specificity and reproducibility. Such tests are easy to standardize and less labour intensive than antibody detection by indirect IFA and thus avoids the requirement of growing SARS-CoV.
RT-PCR has been widely used for the rapid diagnostic of the viral genome in different clinical specimens. Early diagnosis of SARS-CoV infection, which involves viral RNA detection by RT-PCR, first targeted the polymerase (pol) 1b region of the 5’ replicase gene using different formats including one-step or two-step RT-PCR or real-time PCR assays. A comprehensive monitoring of the time periods of RT-PCR diagnosis after disease onset in different types of specimens such as tracheal and nasopharyngeal aspirates, throat swabs, nasal swabs and rectal swabs has also been studies. This study demonstrates that the peak detection rate for SARS-CoV occurred at 2 weeks after the onset of stool or rectal swab specimens and at week 4 for urine specimens [1]. It is likely that the current RT-PCR is not quite sensitive enough to detect the early diagnosis of SARS, showing that the detection rate for probable SARS was only 37.5–50%.
The presence of specific antibodies against various viral components has been a classical diagnostics method. It has been found that anti-NP antibodies in patients’ sera are detected early and with high specificity during the infection. Three different methods, Western blot, ELISA and IFA, used both native and bacterially produced SARS antigens to evaluate serum samples obtained from SARS patients, 40 patients with non-SARS pneumonia, and 38 health individuals. A report indicated that 89% of the SARS patients’ sera were found to be positive to SARS-CoV NP antigen by Western blot that had a strong ability to detect antibodies against SARS. The sensitivity and specificity was reported to be 98.5 and 100% respectively [2]. There was no cross reactivity between the N195 protein and antibodies against chicken, pig and canine coronaviruses. The Western blot assay could distinguish patients with fewer caused by other diseases from that of SARS patients, through reducing the possibility of false positives.
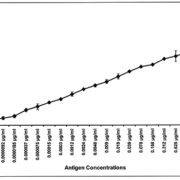
Our earlier study also showed that different combinations of monoclonal antibody (mAb), bispecific antibody (bsmAb), and IgY polyclonal antibody detected the SARS-CoV NP by Immunoswab assay [3] and sandwich ELISA [4] with a sensitivity of 18.5 pg/ml of recombinant SARS-CoV NP antigen in-vitro [Figure 1]. Antibodies against the NP have longer a shelf life and occur in greater abundance in SARS patients than antibodies against other viral components such as the spike protein (SP), membrane and envelope protein. This may be due to the presence of higher levels of NP, compared with other viral proteins, after SARS-CoV infection. A recombinant NP-based IgG ELISA was more sensitive than a recombinant S-protein-based IgG ELISA for diagnosis of SARS-CoV in serum [5–6], due to the highly immunogenic region of N2. It may help in explaining the present results that show less sensitivity of SP detection, compared to a previous NP detection study [4].
Recent studies demonstrate that mAbs and bsmAb could be useful reagents for the diagnosis of SARS-CoV, as well as for functional analysis of SP during infection. Further, the present study shows the development of a novel sandwich ELISA test with a potential use for the diagnosis of SARS-CoV infections based on bsmAb that recognize simultaneously the SP of SARS-CoV and the enzyme peroxidase [7] [Figure 2]. In addition to allowing the rapid diagnosis of SARS infection, the availability of diagnostic tests will help to address important questions such as the period of virus shedding during convalescence, the presence of virus in different body fluids and excreta, and the presence of virus shedding during the incubation period. Until a certain degree of standardization and quality assurance has been achieved for the SARS-CoV laboratory tests, test results must be used with utmost caution in clinical situations. It is strongly advisable to closely check on updated recommendations by the WHO and relevant national organizations regarding the availability and use of such tests.
Limitations
All tests for SARS-CoV available so far have limitations. Extreme caution is therefore necessary when management decisions are to be based on virological test results. In particular, false negative test results (due to low sensitivity, unsuitable sample type, or time of sampling, etc.) may give a false sense of security; in the worst case, they could allow persons carrying the SARS virus, and therefore capable of infecting others, to escape detection.
To aid in the better understanding of SARS, the WHO recommends that sequential samples be stored from patients with suspected or probable SARS – and also close contacts who are not ill themselves – for future use. This is particularly important for the first case(s) recognized in countries that have not previously reported SARS. Data on the clinical and contact history should also be collected in order to obtain a better understanding of the shedding pattern of the virus and the period of transmissibility. Such patient samples should be suitable for viral culture, PCR, antigen detection, immunostaining and/or serological antibody assays. The WHO also encourages each country to designate a reference laboratory for investigation and/or referral of specimens from possible SARS patients.
Future SARS outbreaks
Although the threat of SARS to public health seems to have passed, international health officials continue to remain vigilant. The WHO monitors countries throughout the world for any unusual disease activity (http://www.who.int/csr/sars/en/). Therefore, if another SARS outbreak is to occur, it should be possible to limit the spread of infection using the same measures implemented during the 2002/3 pandemic.
References
1. Chan PK, To WK, Ng KC, Lam RK, Ng TK, et al. Laboratory diagnosis of SARS. Emerg Infect Dis 2004; 10: 825–831.
2. He Q, Chong KH, Chng HH, Leung B, Ling AE, et al. Development of a Western blot assay for detection of antibodies against coronavirus causing severe acute respiratory syndrome. Clin Diagn Lab Immunol 2004; 11: 417–422.
3. Kammila S, Das D, Bhatnagar PK, Sunwoo HH, et al. A rapid point of care immunoswab assay for SARS-CoV detection. J Virol Methods 2008; 152: 77–84.
4. Palaniyappan A, Das D, Kammila S, Suresh MR, Sunwoo HH. Diagnostics of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) nucleocapsid antigen using chicken immunoglobulin Y. Poult Sci 2012; 91: 636–642.
5. Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP. Characterization of novel coronavirus associated with severe acute respiratory syndrome. Science 2003; 300: 1394–1399.
6. Woo PC, Lau SK, Wong BH, Tsoi HW, Fung AM, et al. Differential sensitivities of severe acute respiratory syndrome (SARS) coronavirus spike polypeptide enzyme-linked immunosorbent assay (ELISA) and SARS coronavirus nucleocapsid protein ELISA for serodiagnosis of SARS coronavirus pneumonia. J Clin Microbiol 2005; 43: 3054–3058.
7. Sunwoo HH, Palaniyappan A, Ganguly A, Bhatnagar PK, et al. Quantitative and sensitive detection of the SARS-CoV spike protein using bispecific monoclonal antibody-based enzyme-linked immunoassay. J Virol Methods 2013; 187: 72–78.
The authors
Hoon H. Sunwoo* PhD and Arivazhagan Palaniyappan PhD
Faculty of Pharmacy and Pharmaceutical Sciences,
University of Alberta, Edmonton, Alberta, Canada T6G 2E
*Corresponding author
E-mail: hsunwoo@ualberta.ca