Ensuring high quality viral load results, reduced turnaround times and enhanced laboratory workflows
This article describes the experiences of the Virology Department at Toulouse University Hospital, France, in the evaluation of a new, fully automated molecular diagnostics system for the quantitative determination of nucleic acid targets, such as cytomegalovirus (CMV) DNA and human immunodeficiency virus type 1 (HIV-1) RNA.
by Prof. Jacques Izopet
The 3000-bed Toulouse University Hospital is one of the leading medical facilities in France with a number of research specialties, including immunology and infectious diseases, cardiovascular and metabolic diseases, and oncology. The hospital’s department of biomedical sciences, which employs over 120 medical biologists and 450 engineers and technicians, performs around 6.6 million tests every year. Among these, the department of virology performs a range of culture, serology and molecular biology investigations.
The virology department’s molecular biology laboratory faces a number of challenges in performing viral load analyses for targets, such as cytomegalovirus (CMV) and human immunodeficiency virus type 1 (HIV-1). For CMV, optimal automated quantitative molecular methods are needed to monitor infection, especially among immune-suppressed patients. Similarly, for HIV-1, sensitive biological tools are needed to quantify HIV-1 RNA and to characterize persistent viremia in patients receiving antiretroviral therapy.
These investigations require robust instrumentation and high quality analytical performance. Currently, the laboratory’s viral load measurements are performed using multiple separate instruments for the aliquoting of samples, nucleic acid extraction and amplification/detection. The existing method requires samples to be processed in batches; involves skilled personnel; and is associated with long turnaround times.
Evaluation of a new, fully automated platform
Recently, the laboratory evaluated a new, automated, random access platform for viral load analyses. The DxN VERIS Molecular Diagnostics System (Beckman Coulter) is fully automated from sample entry to result, consolidating DNA or RNA extraction, nucleic acid amplification, quantification and detection onto a single instrument for a number of molecular targets, including CMV, HIV-1, hepatitis B virus (HBV) and hepatitis C virus (HCV).
The aim of the evaluation was to assess the analytical performances of the VERIS CMV assay (for the quantitative determination of CMV DNA in human plasma) and VERIS HIV-1 assay (for the quantitative determination of HIV-1 RNA), and to compare them to the laboratory’s existing method for CMV and HIV-1 (COBAS® AmpliPrep/COBAS® TaqMan® [Roche] coupled to a Hamilton liquid handling system). The laboratory also investigated differences in workflow, comparing the fully automated DxN VERIS System to the existing, semi-automated method.
CMV performance results
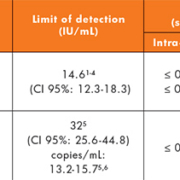
The analytical performance of the VERIS CMV assay system was very good. It demonstrated very high sensitivity and specificity, very good intra/inter-assay reproducibility (both with high viral loads and also when CMV-DNA loads were close to the threshold used to initiate therapy) and a wide analytical range (see Table 1) [1-4].
The clinical performance of the VERIS CMV assay was compared to the laboratory’s existing method for CMV viral load measurement using 169 CMV-positive clinical samples. The two methods were concordant for 88.2% of samples [3,4]. There was good agreement for positive clinical specimens tested by both techniques [1,3,4]. Bland-Altman analysis showed that mean viral loads obtained using the VERIS CMV assay were higher than those obtained using the existing method, with a standard deviation of 0.41 log10IU/mL [4] (Figure 1).
For discordant results, 18/20 (90%) samples tested positive with the VERIS CMV assay and negative with the existing method [3], confirming the very high sensitivity of the VERIS assay [4].
Both assays were also compared for patient monitoring, using four successive samples collected from 17 immunosuppressed patients. This comparison revealed similar trends between the two assays, with overlapping patterns and higher viral loads obtained with the VERIS CMV assay [1,3,4] (Figure 2).
HIV-1 performance results
The VERIS HIV-1 assay demonstrated excellent analytical performance with high sensitivity and specificity, excellent intra/inter-assay reproducibility, and very good linearity across a broad analytical range (table 1) [5,6]. Preliminary data also indicates that there is no influence of HIV-1 subtype on the quantification [6].
The clinical performance of the VERIS HIV-1 assay was assessed using 114 HIV-1 positive samples (mostly HIV-1 subtype B) from Toulouse University Hospital. Passing-Bablok analysis demonstrated that the clinical performance of the VERIS HIV-1 assay correlated well with the existing HIV-1 viral load method, with a small bias for high concentrations (figure 3) [5]. Bland-Altman analysis revealed that the mean difference of HIV-1 RNA concentration obtained using the VERIS HIV-1 assay compared to the existing method was 0.41 log copies/mL (Figure 4) [5,6].
The performance of the two assays was also compared using a panel of 252 HIV-1 positive samples from the Saint-Louis Hospital, Paris, which contained both B (121 samples) and non-B (131 samples) subtypes. Passing-Bablok analysis showed good correlation between the assays, with a small bias for high concentrations (for B and non-B subtypes; for B subtypes only; and for non-B subtypes only) [5]. At very low concentrations (<400 copies/mL), the difference between VERIS and Roche assays was very small (<0.2 log copies/mL) [5].
Workflow efficiencies
The DxN VERIS Molecular Diagnostics System is fully-automated with single sample random access and availability of results as soon as each test is complete (i.e. the first result is available in around 70 minutes for DNA tests and around 100 minutes for RNA tests, with subsequent results every 2.5 minutes). Consolidation of sample extraction and amplification/detection in a single automated platform reduces the number of instruments required for viral load determination from three to just one [5]. It also reduces hands-on time, improving sample security and standardization, and offers a more streamlined workflow [4]. With just 4 steps required for operation (loading of samples onto a rack; placing the rack in the DxN VERIS System; starting the run; reading the auto-verified results), the DxN VERIS System has the potential to revolutionize laboratory practice [7], while the capability to interface with the Laboratory Information System (LIS) ensures ASTM compliance in this respect.
In a workflow analysis for HIV-1 viral load testing at the Toulouse laboratory, the DxN VERIS System was found to reduce complexity of use, with fewer steps (daily maintenance, pre-analytical and post-analytical) and fewer consumables (reduced from >10 to 5) compared to the existing method [5].
The DxN VERIS System also reduced turnaround times for results. The difference in turnaround times between the DxN VERIS System and the existing method was over 25 hours in favour of the DxN VERIS System when the weekend was not taken into account, and over 49 hours in favour of the DxN VERIS System when the weekend was taken into account (figure 5) [5].
Conclusions
In the evaluation at Toulouse University Hospital, the DxN VERIS System demonstrated good analytical and clinical performances in the quantitative determination of CMV DNA and HIV-1 RNA in plasma samples [1-7], comparing well to the laboratory’s existing methodology [1-7] and satisfying quality requirements for the routine monitoring of viral loads in plasma samples [2,4]. It is a completely automated platform, from primary patient sample to result, making it easy-to-use and reliable [1], and offering major improvement in laboratory workflows [5].
The simplified workflow and reduced manual intervention saves staff time, allowing them to focus on other tasks, such as research and innovation [7]. In addition, the single sample random access capabilities of the DxN VERIS System allow laboratories to process samples whenever they are required, without the need for batching, which allows faster results and provides a better service for clinicians and patients [7].
References
1. Mengelle, C, Sauné, K, Haslé, C et al (2014) VERIS/MDx System CMV Assay: a new automated molecular method for quantifying cytomegalovirus-DNA in plasma. Poster presentation, RICAI 2014.
2. Mengelle, C, Sauné, K, Haslé, C (2015) Performance of a completely automated system for monitoring CMV DNA in plasma. Poster presentation, ECCMID, Copenhagen, 2015.
3. Izopet, J, Mengelle, C, Sauné, K (2015) Performance of a new completely automated system for monitoring CMV DNA, HBV DNA, HCV** and HIV** RNA in plasma*. Presented at ECCMID 2015.
4. Mengelle, C, Sandres-Sauné, K, Mansuy, J et al. (2016) Performance of a completely automated system for monitoring CMV DNA in plasma. Journal of Clinical Virology 79: 25–31.
5. Izopet, J (2016) Quantifying HIV-1 RNA with DxN VERIS, a new fully-automated system. Presented at ECCMID 2016.
6. Sauné, K, Haslé, C, Boineau, J (2015) Analytical performance of VERIS MDx system HIV assay for quantifying HIV RNA. Poster presentation, ESCV, Edinburgh, 2015.
7. Izopet, J (2015) Workflow Transformed: A New Fully-automated System for Molecular Diagnostics. Presented at EuroMedLab, Paris , 2015.
The author
Professor Jacques Izopet, Department of Virology, Institut Fédératif de Biologie, CHU Toulouse, France.