ESBL NDP and Carba NP tests: novel techniques for rapid detection of multidrug-resistant bacteria
Two novel biochemical tests, the ESBL NDP and the Carba NP tests, have been recently developed for the early detection of ESBL- or carbapenemase resistance traits in Enterobacteriaceae. Those tests are rapid, sensitive, specific and cost-effective. Implementation of those tests in clinical microbiology laboratories may significantly improve the management and outcome of patients.
by Dr L. Dortet, Dr L. Poirel and Prof. P. Nordmann
Multidrug resistance is now emerging worldwide at an alarming rate among Gram negatives bacteria, causing both community-acquired and nosocomial infections [1–3]. One of the most important emerging resistance traits in Enterobacteriaceae corresponds to the acquisition of resistance to broad-spectrum β-lactams, which is mainly associated with production of clavulanic acid inhibited extended-spectrum β-lactamases (ESBLs) [4, 5]. An ESBL is a β-lactamase that confers reduced susceptibility, i.e. resistance, to the oxyimino-cephalosporins (e.g. cefotaxime, ceftriaxone, ceftazidime) and monobactams (e.g. aztreonam). The hydrolytic activity of ESBLs can be inhibited by several β-lactamase inhibitors such as clavulanic acid and tazobactam. Noteworthy, ESBLs usually do not hydrolyse cephamycins (e.g. cefoxitin and cefotetan) and carbapenems (imipenem, meropenem). In the context of worldwide spread of multidrug resistance, ESBL producers that are mostly Escherichia coli and Klebsiella pneumoniae are not only found as source of hospital-acquired but also of community-acquired infections [4-6]. Consequently, the last line of therapy, carbapenems, is now frequently needed to treat severe infections. However, carbapenem-non-susceptible Enterobacteriaceae due to the production of a carbapenem-hydrolysing enzymes termed carbapenemases, have been reported increasingly [1, 7, 8], leaving us with almost no effective molecules.
Thus, the early detection of ESBL and carbapenemase producers in clinical microbiology is now of utmost importance for determination of appropriate therapeutic schemes and the implementation of infection control measures.
Recently, we have developed two novel tests for rapid identification of (i) ESBL-producing Enterobacteriaceae (ESBL NDP test) [9] and (ii) carbapenemase-producing Enterobacteriaceae and Pseudomonas spp. (Carba NP test) [10–12]. We discuss here the clinical value of those tests.
Detection of ESBLs: Place of the ESBL NDP test in the diagnostic armamentarium
Current techniques for detecting ESBL producers are based on the determination of susceptibility to expanded-spectrum cephalosporins followed by the inhibition of the ESBL activity, mostly by clavulanic acid or tazobactam [13]. The double-disk synergy test, the “E-test” ESBL and the combined disk method have been proposed for that purpose. All those techniques consist of the identification of a synergy between an extended-spectrum generation cephalosporin (ESC) and an inhibitor of β-lactamase (i.e. clavulanic acid or tazobactam) after 18–24h of growth on Mueller-Hinton agar.
This synergy is visualized by (i) a “bouchon de champagne”-shaped image between the extended-spectrum generation cephalosporin and the clavulanate-containing disks for the double-disk synergy test, by (ii) a difference of minimal inhibitory concentration of more than three dilutions between ESC alone and association clavulanate-ESC for the “E-test” ESBL and by (iii) a difference of inhibition diameter of more than 5 mm between an ESC-containing disk and a combined disk containing the same ESC plus clavulanate.
Sensitivities and specificities of the double-disk synergy test and of the E-test are good, ranging from 80 to 95% [13]. However, due to the large diversity of ESBLs [6] that do not hydrolyse ESC similarly, several combinations of those molecules (cefotaxime, ceftazidime and cefepime) together with clavulanate should be tested. Based on the same principle, automated methods for bacterial identification and susceptibility testing are also used in the detection of ESBL-producing organisms. The performance of those systems varies and differs depending on the species investigated with a much higher sensitivity (80–99%) than specificity (50–80%) [13]. However, those tests require mostly overnight growths after isolation of the bacteria, meaning that up to 24–72 h can elapse before ESBL production is detected once the isolate has grown.
Molecular methods (PCR, hybridization, sequencing) based on the detection of ESBL genes have been developed as an alternative. Although classical PCR and DNA arrays necessitate isolation of the bacteria from the clinical sample, real-time PCR based techniques may be performed directly on clinical samples, leading to a decrease of the detection delay. However, these molecular techniques remain costly and require a certain degree of expertise, which is not accessible to non-specialized laboratories. Additionally, those detection methods are able to detect only known genes. They are usually not performed in a routine laboratory but restricted to epidemiological purposes.
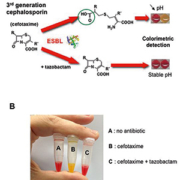
Recently, a rapid and cost-effective biochemical test was developed for the detection of ESBL producers, namely the ESBL NDP test [9]. This test is based on a technique designed to identify the hydrolysis of the β-lactam ring of a cephalosporin (cefotaxime), which generates a carboxyl group, consequently acidifying a medium [Figure 1A]. It can either be performed in a 96-well microtiter plate or into a single tube [Figure 1B]. The acidity resulting from this hydrolysis is identified by the colour change using a pH indicator. Inhibition of ESBL activity is evidenced by adding tazobactam in a complementary well [Figure 1]. The ESBL NDP test may be performed on isolate colonies or directly from clinical samples. When performed on bacterial colonies, the overall sensitivity and specificity of the ESBL NDP test are 92.6% and 100% respectively. The ESBL NDP test can easily differentiate ESBL producers from strains that are resistant to expanded-spectrum cephalosporins by other mechanisms, and from those that are likely to be susceptible to expanded-spectrum cephalosporins. Sensitivity of the test is 100% when the ESBL is of the CTX-M-type. Of note, those CTX-M ESBLs have spread worldwide and have become the most predominant type of ESBL [14]. The ESBL NDP test possesses excellent sensitivity (100%) and specificity (100%) when performed directly from blood cultures. In that case, the gain of time for detection of ESBL producers is ~48h compared to the previously mentioned techniques. Additionally, the ESBL NDP test may also be performed directly on colonies grown on selective media used for the screening of colonized patients, leading to a gain of time of at least 24h for the identification of carriers of ESBL producers and consequently faster implementation of adequate hygiene measures that will further prevent the development of nosocomial outbreaks [2, 5].
Detection of carbapenemases: Place of the Carba NP test in the diagnostic armamentarium
In Enterobacteriaceae, carbapenem resistance may be related either to association of a decrease in bacterial outer-membrane permeability with overexpression of β-lactamases possessing no carbapenemase activity, or to the expression of carbapenemases [7]. The spread of carbapenemase producers is an important clinical issue since carbapenemases confer resistance to most β-lactams. A variety of carbapenemases have been reported, such as Ambler class A carbapenemases of KPC-type, metallo-β-lactamase (Ambler class B) of VIM-, IMP- and NDM-types, and Ambler class D carbapenemase of OXA-48-type [7]. In addition, the detection of carbapenemase producers is a major issue since they are usually associated with many other non-β-lactam resistance determinants, giving rise to multi- or even pandrug-resistant isolates [1, 3].
Potential carbapenemase producers are currently screened first by susceptibility testing based on breakpoint values for carbapenems. Additional non-molecular techniques have been proposed for in vitro identification of carbapenemase production. One of the commonly used techniques corresponds to the modified Hodge test (MHT), which has been used for years. Although the addition of zinc to the culture medium was recently shown to increase the sensitivity of this test [in particular for metallo-β-lactamase (MBL) producers], the MHT remains time-consuming (at least 24h) and may lack of specificity (frequent false-positives with Enterobacter spp. overexpressing their chromosomal cephalosporinase, and false-negatives results with many NDM producers). Other detection methods based on the inhibitory properties of several molecules do exist, either for KPC (e.g. boronic acid, clavulanic acid) or MBL (e.g. EDTA, dipicolinic acid) producers, therefore allowing discrimination between the diverse types of carbapenemases. All those methods are time-consuming since they do require isolation of the bacteria from the infected samples followed by at least an additional 24h period of time for performing the inhibitor-based technique. Several molecular methods such as simplex and multiplex PCRs, DNA hybridization and sequencing are also commonly used for the identification of carbapenemase genes in research laboratories and reference centres. Recently a real-time PCR (RT-PCR) technique has been used for detecting KPC producers directly from blood cultures. Although interesting, this molecular-based technique is costly and requires expertise in molecular techniques.
A rapid and cost-effective biochemical test, the Carba NP test, was recently developed to detect carbapenemase production from isolated colonies [12]. The principle of this test is the same as that of the ESBL NDP test, but uses imipenem as substrate instead (Figure 2A). The Carba NP test differentiates carbapenemase producers (100% sensitivity and 100% specificity) from strains being carbapenem resistant due to non-carbapenemase-mediated mechanisms (Figure 2B) such as combined mechanisms of resistance (outer-membrane permeability defect associated with overproduction of cephalosporinase and/or ESBLs) or from strains that are carbapenem susceptible but express a broad-spectrum β-lactamase without carbapenemase activity (ESBL, plasmid and chromosome-encoded cephalosporinases). Interpretable positive results are always obtained in less than 1h total time, which is unique, making it possible to implement rapid containment measures to limit the spread of carbapenemase producers. The Carba NP test might be performed from colonies recovered from antibiogram (gain of time at least 24h) or from selective media used for screening of carriers (gain of time at least 48h). It was shown to detect carbapenemase producers not only in Enterobacteriaceae [11, 12] but also in Pseudomonas spp. [10]. Additionally, the Carba NP test has also been evaluated for detection of carbapenemase-producing Enterobacteriaceae directly from positive blood cultures [15]. In that case, the Carba NP test has 97.9% sensitivity and 100% specificity. This technique, once applied routinely in clinical laboratories, may guide the first line therapy for treating patients with sepsis, and therefore significantly change the patient outcomes, particularly in areas where carbapenemase producers are highly prevalent (such as Greece, Italy, Turkey, Israel, India). Additionally, when compared to molecular techniques, the Carba NP test may detect any carbapenemase production regardless of the corresponding gene being either known or unknown. Consequently, the Carba NP test is a useful tool for the detection of new carbapenemases that might eventually further disseminate, as recently shown with NDM-1 carbapenemase [8].
Conclusion
The ESBL NDP and the Carba NP tests are rapid, sensitive, specific and cost-effective biochemical tests for the early detection of the most important emerging resistance traits corresponding either to ESBL- or carbapenemase-producing Enterobacteriaceae. Implementation of such tests in the strategies of detection of multidrug-resistant bacteria may significantly improve the management and outcome of colonized and infected patients. Subsequently, the antibiotic stewardship would be improved leading to the decrease of the selective pressure that plays a crucial role in the emergence and spreading of multidrug-resistant bacteria.
Abbreviations
IMP, imipenemase; KPC, Klebsiella pneumoniae carbapenemase; NDM, New Delhi metallo-β-lactamase; VIM, Verona imipenemase; OXA, Oxacillinase
References
1. Schwaber MJ, Carmeli Y. JAMA 2008; 300: 2911–2913.
2. Spellberg B, Blaser M, et al. Clin Infect Dis 2011; 52(Suppl 5): S397–428.
3. Walsh TR, Toleman MA. J Antimicrob Chemother 2011; 67: 1–3.
4. Coque TM, Baquero F, Canton R. Euro Surveill 2008; 13.
5. Pitout JD, Laupland KB. Lancet Infect Dis 2008; 8: 159–166.
6. Poirel L, Bonnin RA, Nordmann P. Infect Genet Evol 2012; 12: 883–893.
7. Nordmann P, Dortet L, Poirel L. Trends Mol Med 2012; 18: 263–272.
8. Nordmann P, Poirel L, et al. Trends Microbiol 2011; 19: 588–595.
9. Nordmann P, Dortet L, Poirel L. J Clin Microbiol 2012; 50: 3016–3022.
10. Dortet L, Poirel L, Nordmann P. J Clin Microbiol 2012; 50: 3773–3776.
11. Dortet L, et al. Antimicrob Agents Chemother 2012; 56: 6437–6440.
12. Nordmann P, Poirel L, Dortet L. Emerg Infect Dis 2012; 18: 1503–1507.
13. Drieux L, Brossier F, et al. Clin Microbiol Infect 2008; 14(Suppl 1): 90–103.
14. Livermore DM, Canton R, et al. J Antimicrob Chemother. 2007; 59: 165–174.
15. Dortet L, Bréchard L, et al. J Antimicrob Chemother (submitted 2012).
The authors
Laurent Dortet, PhD, PharmaD, Laurent Poirel, PhD and
Patrice Nordmann, PhD, MD
Service de Bactériologie-Virologie, INSERM U914 “Emerging Resistance to Antibiotics”, Hôpital de Bicêtre, Assistance Publique/Hôpitaux de Paris, Faculté de Médecine Paris Sud, K.-Bicêtre, France
E-mail: patrice.nordmann@bct.aphp.fr