HCC biomarker, a proteomics approach: the journey from bench to bedside
A goal of clinical proteomics is to find a disease indicator (biomarker) to identify the presence of, or monitor, a disease. It may be surprising that approximately one-third of all cancer cases could be effectively treated if detected at an early enough stage. As a heterogeneous disease, cancer evolves via multiple pathways and is a culmination of a variety of genetic, molecular and clinical events. Given that there is significant variation in the risk of developing cancer and that early detection often results in increased survival, developing technologies capable of identifying patients at highest risk and detecting tumours in the earliest stages of development is a pressing need.
by Dr Gul M. Mustafa, Prof. Cornelis Elferink and Prof. John R. Petersen
Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer, ranking sixth among cancers in incidence worldwide and is the 3rd leading cause of cancer death. Despite some significant improvements in diagnosis and treatment of human liver diseases over the last decade, the HCC mortality rate has not changed to any extent. Currently there are approximately 20,000 new case in the US annually with millions world-wide [1]. The projected rise in the new HCC cases in the US and the world is mainly due to latent hepatitis C virus (HCV) infections in the general population, accounting for approximately 80% of HCC cases several decades after initial infection. The less than 5% survival rate of patients with HCC is primarily due to the disease eluding early detection and diagnosis, when options for effective treatment still remain. Surveillance of patients at highest risk for developing HCC, notably patients with cirrhosis, would benefit greatly from a biomarker assay capable of accurately detecting HCC in its earliest stages when it is still possible to intervene. One of the most widely used markers for HCC is alpha fetoprotein (AFP) although it is non-specific, providing low sensitivity and poor specificity, especially for early detection of HCC [2]. The false-negative rate with AFP level can be 40% for tumours < 3 cm in diameter. More reliable methods such as triple phase Computed Tomography (CT) imaging and liver biopsies exist, but these are expensive and not conducive to long-term surveillance. Therefore, the identification of superior biomarkers will be of huge clinical significance to
at-risk populations.
The ideal biomarker for this type of application would be one where HCC is detected with a high sensitivity and specificity in easily obtained biological samples in a non-invasive, or minimally invasive, manner. Blood represents the best source for detection of HCC related biomarkers, as every cell in the body leaves a record of its physiological state by the products it sheds into the blood, either as a waste or as a signal to neighbouring cells. What some may view as cellular refuse in is reality a diagnostic gold mine. Because of its easy accessibility from patients on a regular basis and because it is in contact with all the tissues in the body, it is an excellent choice for a proteomics approach as it may reveal when changes, such as development of HCC, occur. The systematic analysis of the whole serum or plasma proteome may thus provide a functional meaning to the information provided by genome expression studies. Expression of proteins, their isoforms or post-translational modifications, can be detected by proteomic analysis and these data can provide precious information to better understand the pathologic/molecular basis of HCC [3]. Proteomic analysis may also allow monitoring of the course of the disease process from cirrhosis to HCC, eventually leading to earlier diagnosis which is essential in determining the best course of treatment options and possible outcomes. In addition to earlier diagnosis proteomic analysis may also be useful in measuring the efficacy/progress of treatment or detecting tumour reoccurrence both of which are missing in HCC treatment.
Proteomics analysis
Proteomics analysis is currently considered to be the best tool for the global evaluation of protein expression, and has been widely applied in the analysis of diseases, especially cancer research. For us the approach was to compare the serum/plasma protein profile from patients infected with HCV against the sera from patients with confirmed HCC. Proteins found to be consistently altered between the two patient populations can then be identified and further characterised to determine if they can be used as biomarkers of HCC. While on the surface this sounds simple, due the complexity of the proteome and the wide dynamic concentration range (9 orders of magnitude from pg/mL to mg/mL) of constituent protein/peptide species it is an extremely challenging task. Because the serum/plasma proteome is predominated by high abundance proteins such as albumin and immunoglobulins, extensive fractionation prior to analysis is required. To reduce the few over-represented (i.e. abundant) proteins, without losing any valuable information, existing fractionation methodologies often discard the high abundance carrier proteins, such as albumin, and thus fail to capture the information associated with this valuable resource. We have used aptamer-based technology (Bio-Rad) a technology that reduces the dynamic range and thus retains the complexity of the serum peptidome, in contrast to strategies that just deplete carrier proteins.
Quantitative protein expression profiling
Because proteins entering the blood from surrounding tissue are much less abundant, it is this fraction that is likely to contain most of the undiscovered biomarkers. Quantitative protein expression profiling is a crucial part of proteomics, and such profiling requires methods that are able to efficiently provide accurate and reproducible differential expression values for proteins in two or more biological samples. Thousands of different protein species present in the biological fluid or tissue must be separated, identified and characterised, which cannot be accomplished by a single experimental approach. An effective approach is two-dimensional differential in gel electrophoresis (2D-DIGE) and mass spectrometry [4]. While two-dimensional electrophoresis (2DE) has been widely used for proteomics research, the inter-gel variation along with excessive time/labour costs are major problems. Two-dimensional differential in gel electrophoresis (2D-DIGE) is a modification of 2DE and is considered as one of the most significant advances in quantitative proteomics. Using 2D-DIGE, two samples that are to be compared are pre-labelled with mass- and charge-matched fluorescent cyanine dyes and co-separated on the same 2D gel. The use of internal standards in every gel minimises problems associated with technical variability. Moreover with the great sensitivity and dynamic range that is afforded by the fluorescent dyes, 2D-DIGE can give greater accuracy of quantitation than traditional silver staining. The data captured from these gels using the Imagers, such as the Typhoon trio, along with and proprietary (Decyder) software can be configured to give inter-gel and intra-gel statistical analysis providing both a quantitative and qualitative analysis. We and others are using this approach to identify differentially expressed proteins for differential expression between the pre-cancerous and cancerous patient groups.
Stable isotope labelling
Another technique that can be useful in the analysis of the whole serum proteome is stable isotope labeling using O16/O18. This is a quantitative proteomic technique that distinguishes individual peptides during LC-MS/MS on the basis of a 4 Dalton m/z change after differential O16/O18 labelling that takes place at the C-terminal carboxyl group of tryptic fragments [5]. It is then possible to determine the ratio of individual protein expression levels between the two samples. Alternatively it is possible to use O16/O18 stable isotope labeling to determine the differential expression between two patient groups. In this way the low molecular weight serum peptidome (<20kDa), suspected of harbouring metabolites and degradation products reflecting HCC, can also be interrogated
Selected reaction monitoring
Selected reaction monitoring (SRM), which is used to monitor a precursor and its product ion m/z, is another powerful proteomic tool using tandem mass spectrometry to monitor target peptides within a complex protein digest. The specificity and sensitivity of the approach, as well as its capability to multiplex the measurement of many analytes in parallel, renders it amenable to biomarker discovery and validation proteomics. Using the selectivity of multiple stages of mass selection of tandem mass spectrometers, these targeted SRM assays are the mass spectrometry equivalent of a Western blot. An advantage of using a targeted mass spectrometry-based assay over a traditional Western blot is that it does not rely on the creation of highly selective immunoaffinity reagents. Thus, targeted SRM assays using heavy isotope-labelled internal standards can be multiplexed in quantitative assays that can be directly applicable to clinical settings. A targeted proteomics workflow based on SRM on a triple Quadrupole mass spectrometry platform shows the potential of fast verification of biomarker candidates reducing the gap between discovery and validation in the biomarker pipeline. Although useful, due diligence needs to be exercised in developing and validating SRM assays.
Sample handling
Biomarker research necessitates a clear, rational framework. Technologically, the platform needs to be able to detect low abundant plasma/serum proteins and reproducibly measure them in a high throughput manner. Conceptually, the choice of the technological platform and availability of quality samples should be part of an overall study design that integrates basic and clinical research. Sample preparation is an important and very critical part of clinical proteomics as the collection, sample handling and storage can have a significant impact on the integrity of the proteins being detected. It is so important that a standard operating procedure outlining the steps that should be followed in collecting and storing clinical samples was recently published [6]. In addition to a standardised collection procedure, biological samples need to be carefully chosen based on well-established guidelines either for candidate discovery in the form of controls and the disease being detected or for validation of the candidate biomarkers using well characterised samples.
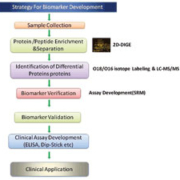
Most importantly, the samples should be representative of the target population and directly address the clinical question. A conceptual structure of a biomarker study can be provided in the form of sequential phases, each having clear objectives and predefined goals [Figure 1]. Furthermore, guidelines for reporting the outcome of biomarker studies are critical to adequately assess the quality of the research, interpretation and generalisation of the results. By being attentive to and applying these considerations, biomarker research should become more efficient and lead to biomarkers that are translatable into the clinical arena.
Aknowledgements
This research was supported by a pilot grant from the Clinical Translational Sciences Award (5UL1RR029876) and the Mary Gibb Jones endowment.
References
1. Kim WR. The burden of hepatitis C in the United States. Hepatology 2002; 36: 30-34.
2. Sterling RK, Wright EC, Morgan TR, Seeff LB, Hoefs JC, Di Bisceglie AM, Dienstag JL, Lok AS. Frequency of elevated hepatocellular carcinoma (HCC) biomarkers in patients with advanced hepatitis C. Am J Gastroenterol 2012; 107(1): 64-74.
3. Maria P, Laura ML, Antonio RA, Jose LM, Javier B, Ruben C, Jordi M and Manuel de la Mata. Proteomic analysis for developing new biomarkers of hepatocellular carcinoma. World J Hepatol 2010; 2(3): 127-135.
4. Sun W, Xing B, Sun Y, Du X, Lu M, Hao C, Lu Z, Mi W, Wu S, Wei H, Gao X, Zhu Y, Jiang Y, Qian X, He F. Proteome analysis of hepatocellular carcinoma by two-dimensional difference gel electrophoresis: novel protein markers in hepatocellular carcinoma tissues. Mol Cell Proteomics 2007; 6(10): 1798-808.
5. Miyagi M, Rao KC. Proteolytic 18O-labeling strategies for quantitative proteomics. Mass Spectrom Rev 2007; 26(1):121-36.
6. Tuck MK et al. Standard operating procedures for serum and plasma collection: early detection research network consensus statement standard operating procedure integration working group. J Proteome Res 2009; 1: 113-117.
The authors
Gul M. Mustafa, Ph.D. Postdoctoral Fellow, Department of Pharmacology
Cornelis Elferink, Ph.D., Professor, Department of Pharmacology, Director Sealy Center Environmental Health and Medicine
John R. Petersen, Ph.D., Professor and Director Victory Lakes Clinical Laboratory, Department of Pathology,
University of Texas Medical Branch
301 University Boulevard
Galveston, Texas 77555, USA
e-mail: jrpeters@utmb.edu