Image analysis enables standardized and quantitative pathology
The analysis of histopathology slides is routinely performed in a manual, semi-quantitative manner which is open to observer variability. This article summarizes how technological advances in image analysis software allow the objective and standardized quantification of such samples while driving pathology towards a more personalized medicine.
by Dr Peter Caie
Introduction
The assessment of stained tissue sections by manual observation down a microscope has been, and still is, the steadfast manner in which histopathologists observe diseased tissue architecture in order to report on a patient’s prognosis. The tissue, for example the tumour microenvironment, is complex, highly heterogeneous and heterotypic. Although specific stains exist to aid in the identification and semi-quantification of histopathological features or biomarkers, the empirical field is subjective and therefore open to observer variability. In colorectal cancer (CRC) this can be the case for reporting items from the minimal core clinical data set such as differentiation [1] or promising histopathological features such as tumour budding [2] and lymphovascular invasion [3]. Similarly, in breast cancer discrepancies exist in the reproducibilty of manual reporting of human epidermal receptor protein-2 (HER2) by fluorescence in situ hybridization (FISH) or immunohistochemistry and the scoring of estrogen receptor (ER), both of which have predictive implications for patient treatment strategies [4]. Some reproducibility issues may be overcome through molecular pathology and the objective automated quantification of molecular biomarkers extracted from patient tissue samples. Modern methodology in quantitative pathology, spanning the classical ‘omics’ fields, has the ability to create a wealth of complex big data. Indeed, the field of molecular pathology has seen an explosion of big data specifically in translational genomics, transcriptomics and proteomics and which has the ability to map aberrant molecular pathways with direct impact on clinical decisions. The automated and standardized extraction of large data sets from tissue, has been termed ‘tissue datafication’. The automated quantification of molecular pathology, such as next-generation sequencing (NCS), gene-chip transcriptomics and reverse phase protein arrays may still suffer from reproducibility issues. These may occur from poor and small sample sizes or tissue artefacts which can stem from multiple sources: surgical ischemia, fixation and sample preparation. Standardization is therefore the key to accurate tissue datafication in order to report reproducible results which translate to the clinic. Tissue heterogeneity, both inter-patient and intra-patient, poses a very real problem for the effective personalized treatment decisions for patients. Tissue is often homogenized in order to extract the DNA, RNA or protein required for many molecular pathology techniques. In doing so the tissue heterogeneity (both subpopulation and spatial heterogeneity) is invariably lost and a single end-point is reported from the most dominant signal within the complex sample. A patient may therefore initially respond to a targeted treatment such as cetuximab in CRC but relapse within a set time period because of the existence of resistant KRAS and BRAF mutated subpopulations within the tumour [5]. Effective personalized combination therapy must rely on the capture of molecular end-points across the heterogeneous disease. Quantitative pathology must take into account the imperfection of the tissue sample as well as its heterogeneity in order to produce standardized and reproducible results. With the advent of digital pathology and associated image analysis solutions, histopathology has joined the ranks of molecular pathology with the ability to generate robust and standardized quantitative big data. Image analysis can also capture the heterogeneity across a patient sample by digitally segmenting the tumour subpopulations while extracting quantitative hierarchical morphological or biomarker data (Fig. 1). This review will discuss datafication of the tissue section through image analysis and its benefits as well as some of the challenges within the field.
Quantitative pathology through image analysis
Image analysis has been well established in order to quantify in vitro cell-based assays [6, 7] but has been slow to translate to molecular pathology and histopathology. This is in part due to the more complex and heterogeneous nature of the tissue as well as the need for extensive validation for clinical research compared with cell culture work. Advances in both whole-slide scanners and analysis software are now making the translation of image analysis to clinical research a reality. The use of standardized and automated image analysis solutions overcomes the reproducibility issues associated with manual semi-quantitative scoring of tissue as it negates observer variability. Image analysis has many uses within quantitative histopathology where it can report biomarker expression at sub-cellular resolution, quantify set histopathological features, identify heterogeneous subpopulations or the spatial heterogeneity of tumour and host interaction as well as identify novel histopathological features. Standardization is always the key to reproducible results and the field of image analysis is no different. Standardization and validation must be present throughout the entire process from tissue section cutting, mounting, labelling and digitizing. There are a growing number of whole-slide imagers on the market but it is paramount that these allow the use of identical image capture profiles and associated image quality across all the patient samples used in a study. Once the tissue is digitized in a standardized manner the image analysis algorithms themselves must be of a high enough quality in order to deal with the complex and heterogeneous tissue. Simplified algorithms have their use for basic biomarker quantification but may report false results or classifications owing to heterogeneous cell populations or inter-patient heterogeneity. Autofluorescence or non-specific staining in the sample may result in the reporting of false positives or inaccurate parameters when quantifying histopathological features in the complex tumour microenvironment. The image analysis workflow must therefore be robust enough to take into account or build in quality control steps to negate tissue labelling artefact [8].
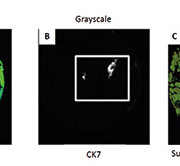
Image analysis can quantify biomarkers
Whole-slide image analysis of molecular biomarkers labelled via antibodies or probes such as in FISH, avoids the contamination of signals from heterogeneous subpopulations that occur when the tissue is homogenized (Fig. 2A). This has advantages over destructive assays as the tissue structure, spatial orientation and sub-localization of molecules are retained [9] and heterogeneity can be compartmentalized and quantified while providing insight into cellular interactions within the tumour and its microenvironment. In order to quantify the biomarker in question the algorithm must segment the cells and nuclei within a region of interest, e.g. the tumour or stroma (Fig. 2B). This gives a further advantage to automated image analysis as morphometric and texture parameters may be captured and co-registered to the cell’s expression of the desired biomarker. This additional information can be used to identify a morphological surrogate to a biomarker or to capture a more definitive result that reduces false positives. When immunofluorescence is applied to biomarker quantification a continuous data capture across the dynamic range of intensity can be reported. The intensity of the fluorophore signal directly correlates to the level of protein expression and therefore returns a more accurate result than the classical 1+, 2+, 3+ manual scoring of chromogenic assays. This continuous data can be used to calculate robust cut-off points for positive and negative expression, or for patient categorization, in software such as X-Tile[ 10] or TMA Navigator [11].
Image analysis can quantify histopathological features
Image analysis may also be employed for the quantification of histopathological features. Observer variability occurs when manual semi-quantification of certain set histopathological features across tissue sections stained with hematoxylin and eosin (H&E) are reported [1–3]. Automated image analysis with the aid of specific labels negates observer variability and introduces standardization which is applicable across heterogeneous patient cohorts. In this manner tumour buds, lymphatic vessel density and invasion were co-registered upon the same tissue section and all quantified using the same algorithm across a CRC patient cohort [8]. This methodology allowed the computer-based algorithm to quantify small lymphatic vessels that were invaded by up to five cancer cells and which often go unreported because of their obscurity in H&E stained sections (Fig. 3). The results showed that these so called ‘occult lymphatic invasion’ events were independently predictive of poor prognosis in stage II CRC patients.
Similarly image analysis may be employed to quantify the host response to the tumour and not just the tumour itself; such as the lymphocytic infiltration within the cancer microenvironment. The immunoscore in CRC uses image analysis to quantify CD3+ and CD8+ lymphocytes at either the invasive front or the centre of the tumour section [12]. The automated quantification of lymphocytes and their spatial heterogeneity have also been shown to be prognostic in breast cancer [13].
Image analysis can identify novel features
Research pathologists apply their extensive experience to identify novel or significant prognostic features within the tissue section. Automated segmentation of digitized tissue sections now allows the quantification and standardization of complex and subtle morphological features or signatures in a continuous data capture manner. These features are extracted from every possible computer segmented object within the image. This image analysis methodology quantifies and profiles the complex phenome of the tumour’s microenvironment in an a priori ‘measure-everything big-data’ approach. Parameters extracted from single objects segmented across the digitized tissue section include morphometrics, texture and spatial heterogeneity. This is performed in an attempt to identify and quantify novel clinically relevant histopathological objects or predictive features from large exported image based multi-parametric big data sets. This emerging methodology has been termed ‘Tissue Phenomics’ by Gerd Binnig a Nobel Laureate and expert in image analysis. These objects may represent single or combinations of morphometrically quantifiable histological features, which may prove too subtle to observe by eye but which could prove prognostic or predictive. Beck et al. demonstrated this technique in breast cancer and found the stromal microenvironment to be specifically relevant to prognosis [14]. The big data created by image analysis approaches such as these needs to be distilled in order to identify the significant parameters which answer the clinical question being investigated. Bioinformatics must be applied which allows redundant parameters to be discarded and clinically relevant cut-offs to be applied to the remaining significant features. The reduced end result of a few significant parameters from potentially thousands of captured features should form a clinically translatable test which must then be validated across multiple international cohorts.
Future developments and challenges to the field
Technological advances in both image capture and analysis are beginning to see the translational of automated big data from the realm of academic research to clinical tests. Further technological advances such as co-registering of tissue sections and the ability to multiplex numerous biomarkers on a single tissue section will add greater value to the field. This multiplexed, next-generation immunohistochemistry [15] approach coupled with automated quantification may allow whole molecular pathways to be mapped at the single cell level. There are, however, challenges within the field. The automated quantification of pathology requires expensive whole-slide scanners as well as image analysis workstations alongside associated IT infrastructure to archive and keep secure the images and associated analysis. Fast Ethernet connections are also essential to recall these images in a time dependent manner. Another challenge is the acceptance of automated analysis within the clinical environment. This challenge will need to be overcome by validating the standardized and automated image analysis algorithms across multiple cohorts. The many applications of the field, such as objective, standardized and reproducible quantification of biomarkers, histopathological features and the profiling of a tumour’s heterogeneity hold advantages for both the pathologist and the patient. The negating of observer variability should increase the accuracy of patient results as should the application of clinically relevant categorical cut-offs across a continuous data set captured per patient. The capture of the molecular and histopathological prognostic and predictive signatures across heterogeneous subpopulations as the potential to turn traditional population based statistics into a more personalized one which informs the optimal treatment regimen for the individual patient.
References
1. Compton CC. Colorectal carcinoma: diagnostic, prognostic, and molecular features. Mod Pathol. 2003; 16: 376–388.
2. Puppa G, Senore C, Sheahan K, Vieth M, et al. Diagnostic reproducibility of tumour budding in colorectal cancer: a multicentre, multinational study using virtual microscopy. Histopathology 2012; 61: 562–575.
3. Harris EI, Lewin DN, Wang HL, Lauwers GY, et al. Lymphovascular invasion in colorectal cancer: an interobserver variability study. Am J Surg Pathol. 2008; 32:1816–1821.
4. Gown AM. Current issues in ER and HER2 testing by IHC in breast cancer. Mod Pathol. 2008; 21: S8–S15.
5. Baldus SE, Schaefer KL, Engers R, Hartleb D, et al. Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res. 2010; 16: 790–799.
6. Caie PD, Walls RE, Ingleston-Orme A, Daya S, et al. High-content phenotypic profiling of drug response signatures across distinct cancer cells. Mol Cancer Ther. 2010; 9: 1913–1926.
7. Gasparri F, Mariani M, Sola F, Galvani A. Quantification of the proliferation index of human dermal fibroblast cultures with the ArrayScan high-content screening reader. J Biomol Screen. 2004; 9: 232–243.
8. Caie PD, Turnbull AK, Farrington SM, Oniscu A, Harrison DJ. Quantification of tumour budding, lymphatic vessel density and invasion through image analysis in colorectal cancer. J Transl Med. 2014; 12: 156.
9. Kumar A, Rao A, Bhavani S, Newberg JY, Murphy RF. Automated analysis of immunohistochemistry images identifies candidate location biomarkers for cancers. Proc Natl Acad Sci U S A 2014; 111: 18249–18254.
10. Camp RL, Dolled-Filhart M, Rimm CL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004; 10: 7252–7259.
11. Lubbock AL, Katz E, Harrison DJ, Overton IM. TMA Navigator: Network inference, patient stratification and survival analysis with tissue microarray data. Nucleic Acids Res. 2013; 41(Web Server issue): W562–568.
12. Galon J, Mlecnik B, Bindea G, Angell HK, et al. Towards the introduction of the Immunoscore in the classification of malignant tumors. J Pathol. 2013; 232: 199–209.
13. Yuan Y. Modelling the spatial heterogeneity and molecular correlates of lymphocytic infiltration in triple-negative breast cancer. J R Soc Interface 2015; 12: 20141153.
14. Beck AH, Sangoi AR, Leung S, Marinelli RJ, et al. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med. 2011; 3: 108ra113.
15. Rimm DL. Next-gen immunohistochemistry. Nat Methods 2014; 11: 381–383.
The author
Peter Caie PhD
School of Medicine, University of St Andrews, St Andrews KY16 9TF, UK
E-mail: Pdc5@st-andrews.ac.uk