MicroRNAs: New tools to tackle liver cancer progression
Primary hepatic tumours are one of the most aggressive and resistant forms of cancer. Early diagnosis of liver cancer and the development of more accurate markers for biological classification are crucial to improving the clinical management and survival of patients. This article discusses the emerging use of microRNAs for the diagnosis of liver cancer.
by Dr Luc Gailhouste and Dr Takahiro Ochiya
Liver cancer and diagnosis
Primary liver cancer is mainly represented by hepatocellular carcinoma (HCC) and accounts for almost 90% of primitive hepatic malignancies. Statistically, HCC is the third most common cause of death from cancer worldwide [1] and is generally encountered in patients exhibiting an underlying chronic liver disease such as hepatitis B virus (HBV) and/or C virus (HCV) infection, alcohol abuse, or liver steatosis. Chronic hepatitis leads to fibrosis and gradually evolves into cirrhosis. Global studies estimate that approximately 80–90% of all HCCs arise from cirrhotic livers. Despite great advances in the treatment of the disease, hepatic cancer exhibits one of the lowest remission rates (less than 10% after five years), mainly due to its late diagnosis and high resistance to the conventional agents of chemotherapy. Indeed, as such a disease tends to remain asymptomatic, approximately 50% of newly diagnosed patients already exhibit late advancement.
Common HCC diagnostic methods include liver imaging techniques such as triphasic computed tomography scanning, magnetic resonance imaging (MRI), and abdominal ultrasound [2]. A panel of serological biochemical markers, including aminotransferases ALAT and ASAT, has also been used for several decades to monitor liver pathologies in a non-invasive manner.
Until recently, imaging tests were frequently combined with the non-invasive measurement of serum alpha-fetoprotein (AFP). Normally produced by the fetal liver, AFP decreases soon after birth whereas its high level in adults can be correlated with the appearance of malignant hepatic disease. However, the American Association for the Study of Liver Diseases (AASLD), in its practice guidelines, discontinued the use of the blood tumour marker AFP for surveillance and diagnosis due to the limited sensitivity and specificity of the method. When uncertainty regarding the diagnosis persists, a percutaneous biopsy followed by histological examination of the nodule is indicated [3]. This technique remains the gold standard method for determining the degree of underlying fibrosis and shows appreciable sensitivity (more than 80%) for HCC diagnosis.
An important breakthrough in the clinical management of liver cancer would come from the accurate correlation of the alterations of cancer-related genes and the tumour phenotype. Although HCC lesions can be broadly distinguished by histological or immunological assessment, their prognosis and clinical evolution vary greatly from one individual to another. The discovery of innovative and effective biomarkers ensuring an early diagnosis of the disease correlated with the etiology, the pathogenic tendency, and the malignancy of the tumour could significantly enhance the molecular assessment of HCC and its classification in order to maximize the positive response of therapeutics.
MicroRNAs: biogenesis and mechanism of action
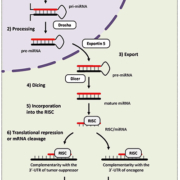
MicroRNAs (miRNAs) constitute a group of evolutionary conserved small non-coding RNAs of approximately 22 nucleotides that accurately regulate gene expression by complementary base pairing with the 3’-untranslated regions (3’-UTRs) of messenger RNAs (mRNAs) [4]. These post-transcriptional regulators were first evidenced in C. elegans by Ambros and co-workers who discovered that lin-4, a gene known to control the timing of nematode larval development, did not code for a protein but produced small RNAs that specifically bind to lin-14 mRNA and repress its translation.
miRNA biogenesis is a multistep process that has been reviewed extensively [Figure 1]. An essential feature of miRNAs is that a single miRNA can recognize numerous mRNAs, and, conversely, one mRNA can be recognized by several miRNAs. These pleiotropic properties enable miRNAs to exert wide control over a plethora of targets, attesting to the complexity of this mechanism of gene expression regulation. Several reports have described the key role of these post-transcriptional regulators in the control of diverse biological processes such as development, differentiation, cell proliferation, and apoptosis. The alterations of miRNA expression have also been reported in a wide range of human diseases, including cancer [5].
In HCC, the atypical expression of miRNAs frequently contributes to the deregulation of critical genes known to play an essential role in tumorigenesis and cancer progression. The current consensus is that cancer-related miRNAs function as oncogenes or tumour suppressors [6]. As for other malignancies, two situations can occur in HCC: (i) tumour suppressor miRNAs can be downregulated in liver cancer and cause the upregulation of oncogenic target genes repressed in normal hepatic tissues, increasing cell growth, invasion abilities, or drug resistance; (ii) oncogenic miRNAs, also called oncomirs, can be upregulated in HCC and can downregulate their target tumour suppressor genes, potentially leading to hepatocarcinogenesis.
miRNA as a diagnostic tool
As miRNA signatures are believed to serve as accurate molecular biomarkers for the clinical classification of HCC tumours, the availability of consistent technologies that enable the detection of miRNAs has become of interest for both fundamental and clinical purposes. The most current detection methods commonly used are microarray and real-time quantitative polymerase chain reaction (RT-qPCR).
Microarray analysis presents the advantage of offering a high speed of screening by employing various miRNA probes within a single microchip. However, the technique has lower sensitivity and specificity than RT-qPCR, which is the most widely used method.
miRNA RT-qPCR is based on the use of stem–loop primers, which can specifically bind to the mature miRNA during reverse transcription, granting a high degree of accuracy to the method [7]. Analysis of miRNAs by RT-qPCR is a cost-effective technique and, due to its efficiency, a valuable way to validate miRNA signatures. Moreover, the development of RT-qPCR protocols has improved the sensitivity of miRNA detection down to a few nanograms of total RNAs. This amount can be easily and routinely obtained by extracting total RNAs from a small fragment of a hepatic percutaneous biopsy.
A plethora of studies have already reported various miRNA profiles potentially reflecting HCC initiation and progression that could be employed as specific cancer biomarkers [8]. Comparative analysis of bibliographic data provides evidence of the persistent augmentation of miR-21 in cancer, regardless of the tumour origin. In the HCC, miR-21 is also frequently overexpressed where it acts as an oncogenic miRNA. The major overexpression of miR-21 is associated with the inhibition of the tumour suppressor PTEN and the poor differentiation of the tumour. The use of an miRNA-based classification correlated with the etiology and the aggressiveness of the tumour appears very promising, as it could significantly enhance the accuracy of the molecular diagnosis of HCC and its classification, leading to the consideration of more appropriate therapeutic strategies.
In this regard, Budhu and collaborators defined a combination of 20 miRNAs as an HCC metastasis signature and showed that this 20-miRNA-based profile was capable of predicting the survival and recurrence of HCC in patients with multinodular or single tumours, including those at an early stage of the disease [9]. Remarkably, the highlighted expression profile showed a similar accuracy regarding patient prognosis when compared to the conventional clinical parameters, suggesting the relevance of this miRNA signature. Consequently, the profiling of aberrantly expressed cancer-related miRNAs might establish the basis for the development of a rational system of classification in order to refine the diagnosis and the prediction of HCC evolution.
Tumour suppressor miRNA: the case of miR-122
The case of miR-122 is of prime interest, first, because it represents by itself more than half of the total amount of miRNAs expressed in the liver [10]. Remarkably, miR-122 is a key host factor required for HCV replication. A phase 2 clinical trial was recently initiated that reported the world’s first miRNA-based therapy targeting miR-122 in HCV-infected patients using the locked nucleic acid (LNA)-modified antisense oligonucleotide miravirsen [11]. Thus, a four-week miravirsen treatment by subcutaneous injection provided long-lasting antiviral activity and was well tolerated.
However, the experimental silencing of miR-122 resulted in increased expression of hundreds of genes normally repressed in normal hepatocytes. The miR-122 knockout mouse model displays hepatosteatosis, fibrosis, and a high incidence of HCC, suggesting the tumour suppressor role of miR-122 in the liver. In primary liver carcinoma, the existence of an inverse correlation was demonstrated between the expression of miR-122 and cyclin G1, which is highly implicated in cell cycle progression.
Regarding the potential of miR-122 as a diagnostic biomarker in liver cancer, numerous studies have already reported the significant and specific downregulation of miR-122 expression in both human and rodent HCC models. Obviously, miR-122 was shown as downregulated in more than 70% of the samples obtained from HCC patients with underlying cirrhosis as well as in 100% of the HCC-derived cell lines [12].
To illustrate this statement, we analyzed the expression levels of miR-122 in 20 patients who exhibited HCC using RT-qPCR. Following RNA extraction from biopsies with the miRNeasy Mini Kit (Qiagen), 100 ng of total RNA was reverse-transcribed using the Taqman miRNA Reverse Transcription Kit (Applied Biosystems). The expression levels of mature miR-122 were determined in each sample by RT-qPCR with Taqman Universal PCR Master Mix in a 7300 Real-Time PCR System from Applied Biosystems. The expression levels of miRNAs were normalized with respect to the endogenous levels of RNU6B. RT-qPCR data were obtained easily and rapidly by a routinely conventional method used in our laboratory. As a result, miR-122 expression was reduced more than threefold in HCC biopsies relative to the normal liver group (median 0.935 and 3.495, respectively; P<0.0001, Mann–Whitney U test) [Figure 2]. These data suggest that cancer-related miRNAs, such as miR-122, which are deregulated in HCC tissues, could be relevant with regard to the development of new diagnostic tools and the clinical management of liver cancer patients.
Conclusions and emerging approaches
The expression profile of specific miRNAs has been found to reflect the biological behaviour of HCC tumours, such as aggressiveness, invasiveness, or drug resistance. As a consequence, miRNA investigations may offer opportunities to determine miRNA signatures that would provide valuable information to stratify and refine HCC diagnosis in terms of prognosis, response to treatment, and disease relapse. Recently, tumour-derived miRNAs have been efficiently detected in the serum of patients and characterized as potential non-invasive biomarkers for HCC.
The concept that miRNAs could serve as potential plasma markers for liver diseases is, thus, gaining attention. Due to its frequent deregulation in viral hepatitis, cirrhosis, and cancer as well as its specific and massive expression in the liver, the assessment of serum miR-122 could represent one reliable strategy for the non-invasive diagnosis of chronic liver pathologies. Although the process of assessing serum miRNAs remains under improvement, cancer-related circulating miRNAs represent an exciting and promising field of investigation for the development of more accurate technologies for the early diagnosis of HCC.
References
1. Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer 2006; 6: 674–687.
2. Befeler AS, Di Bisceglie AM. Hepatocellular carcinoma: diagnosis and treatment. Gastroenterology 2002; 122: 1609–1619.
3. Ryder SD. Guidelines for the diagnosis and treatment of hepatocellular carcinoma (HCC) in adults. Gut 2003; 52(Suppl 3): iii1–8.
4. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297.
5. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006; 6: 857–866.
6. Gailhouste L, Ochiya T. Cancer-related microRNAs and their role as tumor suppressors and oncogenes in hepatocellular carcinoma. Histol Histopathol 2012.
7. Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 2005; 33: e179.
8. Gailhouste L, Gomez-Santos L, Ochiya T. Potential applications of miRNAs as diagnostic and prognostic markers in liver cancer. Front Biosci 2013; 18: 199–223.
9. Budhu A, Jia HL, Forgues M, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology 2008; 47: 897–907.
10. Girard M, Jacquemin E, Munnich A, et al. miR-122, a paradigm for the role of microRNAs in the liver. J Hepatol 2008; 48: 648–656.
11. Lindow M, Kauppinen S. Discovering the first microRNA-targeted drug. J Cell Biol 2012; 199: 407–412.
12. Gramantieri L, Ferracin M, Fornari F, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res 2007; 67: 6092–6099.
The authors
Luc Gailhouste PhD and
Takahiro Ochiya PhD
Division of Molecular and Cellular
Medicine, National Cancer Center Research Institute, Tokyo, Japan