Molecular detection and quantification of human rhinoviruses
Introduction
Human rhinoviruses (HRV) are small, positive-sense RNA viruses within the family Picornaviridae. Over 150 genotypes of this important human pathogen have been recognized within species HRVA, HRVB, and HRVC of the genus Enterovirus (http//:www.picornaviridae.com). HRV infections occur throughout the year and throughout the world. HRV are responsible for a high incidence and wide range of respiratory infections in all populations, including one-half to two-thirds of all common colds and many cases of otitis media and sinusitis in the upper respiratory tract. Lower tract infections include bronchiolitis, pneumonia and exacerbation of disease in children with asthma and cystic fibrosis, and in adults with chronic obstructive pulmonary disease. Cases of serious illness and even death due to HRV have been reported, especially in immunocompromised individuals, the elderly and infants [1, 2].
Laboratory detection of HRV is important for diagnosis and surveillance, especially in high risk populations. HRV are frequently detected as co-infections with other respiratory viruses and in individuals with long-term and asymptomatic shedding [3]. In addition to qualitative detection, accurate quantification of HRV RNA in clinical samples is needed for studies on the association of HRV viral load with viral transmission and with patient symptoms and outcomes. Viral-load studies of other respiratory viruses have shown that a correlation exists between quantity of virus and disease severity. HRV viral-load determinations may also be important for patient management, especially in asymptomatic patients who test positive for HRV at low levels. More importantly, accurate HRV viral-load assessments will be necessary for evaluating the performance of potential HRV antiviral drugs [4].
Detection
HRV were initially detected by growth in cell culture. Approximately 100 serotypes of HRV grown in cell culture were antigenically characterized by their reactions with various antisera. The serotypes were subsequently classified into two groups, A and B, according to their sensitivity towards antivirus agents [5] and are now included in HRV species A (80 genotypes) and B (32 genotypes) based on genetic sequencing. Cell culture is sensitive for detection of many, but not all HRV genotypes; 55 HRV that do not grow in the cell culture lines normally used in the clinical laboratory and have been detected only by molecular methods are classified in HRV species C (http//:www.picornaviridae.com).
The use of molecular methods for the detection of HRV in clinical specimens has provided more accurate information about the disease burden and epidemiology of these ubiquitous viruses. The molecular method most often used to detect HRV is real-time reverse-transcription (RT)-PCR [3]. RT-PCR assays, when accompanied by amplification of serially diluted standards of known RNA copy numbers (RT-qPCR), can be used to quantify the number of viral copies in a sample. By comparing the PCR Ct value (the PCR cycle at which fluorescence reaches a certain threshold) of a clinical specimen to the standard curve, the relative quantity of the analyte can be calculated [6].
Within the HRV genome, the region most frequently targeted for RT-PCR by clinical assays is the 5’ non-coding region (NCR), which exhibits the most sequence homology among the HRV genotypes. However, even in this region, there is a lot of sequence diversity, which makes it challenging to design a single, consensus PCR primer and probe set to amplify all HRV genotypes with equal efficiency. In order to amplify HRV genotypes with diverse sequences in the prime/probe binding regions, consensus PCR primer and probe sets have been designed with degenerate and modified bases or multiple oligonucleotides [7–10]. However, consensus RT-qPCR assays may not give accurate quantitative results for all HRV genotypes due to amplification inefficiency caused by base mismatches between the consensus primers and probe and the viral sequences [11].
Quantitation by RT-qPCR
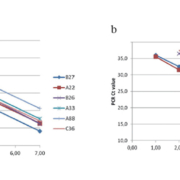
To determine if a consensus RT-qPCR assay [7] could be used to accurately quantify all genotypes of HRV, including those with sequence differences in the primer and probe binding regions, we compared the efficiency and sensitivity of a consensus RT-qPCR assay to that of genotype-specific RT-qPCR assays [4]. In Figure 1(a), the results of RT-qPCR assays using type-specific primers and probes, which exactly match the target sequences, show standard curves indicating accurate and sensitive quantification of RNA transcripts from six specific HRV genotypes. However, RT-qPCR using a consensus HRV primer and probe set did not give accurate or sensitive quantification for some HRV genotypes, especially types A33 and A88 (Fig. 1b). RNA from HRV genotypes with base mismatches between the consensus primer and probe sequences and the specific viral sequences was inaccurately quantified using the consensus assay, most likely due to poor amplification efficiency.
Quantitation by RT-dPCR
Digital RT-PCR (RT-dPCR), which provides absolute nucleic acid quantification without the need for PCR Ct values and standard curves and is less affected by poor amplification efficiency, may perform better than RT-qPCR for quantification of HRV RNA. In dPCR, an amplification reaction, which contains fluorescent dye to measure amplification, is divided into 12?000 to 200?000 independent partitions, each ideally containing no more than one target molecule. The reaction is amplified to end point and the number of fluorescent (positive) and non-fluorescent (negative) partitions is counted. In specimens with more targets than partitions, Poisson statistics are used to calculate the average number of targets per positive partition and thus, the number of targets in the original sample [12, 13]. Compared to qPCR, dPCR is less susceptible to amplification inefficiency caused by primer/probe sequence mismatches because quantification derives from a PCR reaction that cycles to endpoint rather than from an amplification curve as in qPCR. Accurate quantification by dPCR is also not dependent on a well-calibrated standard [14]. These characteristics make dPCR especially useful for quantifying viral targets with many subtypes and high sequence diversity that leads to mismatches between targets and PCR primer and probe sequences, such as HRV.
To determine if consensus RT-dPCR would perform better than consensus RT-qPCR for quantification of HRV genotypes, we similarly tested RNA transcripts of HRV genotypes, including some with sequence variation in the consensus primer and probe binding region, by RT-dPCR using both type-specific and consensus primers and probes. In Figure 2(a), the results of RT-dPCR assays using type-specific primers and probes show good correlations between the expected number of RNA copies/reaction and the observed number. When amplified by RT-dPCR using the consensus assay (Fig. 1b), in contrast to RT-qPCR, the observed number of RNA copies/reaction was also closely correlated with the expected number for most of the HRV genotypes tested.
In a previous study [4], data from 16 HRV genotypes that represented the consensus primer and probe binding sequences of 128 genotypes indicated that, when using consensus primers and probe, RT-dPCR quantification of HRV RNA was more accurate than that of RT-qPCR for some genotypes. We found that although the consensus RT-qPCR did accurately quantify many HRV genotypes, it did not accurately quantify all genotypes of HRV due to sub-optimal amplification of genotypes with sequences that do not exactly match those of the primers and probe. Consensus RT-dPCR, however, did not overcome all sequence mismatch-induced amplification inefficiency, as evidenced by genotype A88 (Fig. 2b), which has a single mismatch near the middle of the forward primer.
Although RT-dPCR has been shown to be more accurate than RT-qPCR for quantification of HRV and may be applicable to other viruses with high sequence diversity, like HIV and HBV, it has some disadvantages for routine use in a clinical laboratory. RT-dPCR has a more limited dynamic range compared to RT-qPCR (104 for RT-qPCR compared to 108 for RT-qPCR), which would require dilution and retesting of samples with high viral loads. Running an RT-dPCR assay requires more hands-on technician time and has a lower throughput than current RT-qPCR assays. Digital PCR instruments and reagents are also currently more expensive than most qPCR systems.
Conclusion
In conclusion, dPCR was a better alternative to qPCR on RNA templates known to have significant sequence diversity that cannot be avoided during primer and probe design and should be considered the better molecular method for quantification of HRV in respiratory specimens.
References
1. Brownlee JW, Turner RB. New developments in the epidemiology and clinical spectrum of rhinovirus infections. Curr Opin Pediatr 2008: 20: 67–71.
2. Gern JE. The ABCs of rhinoviruses, wheezing, and asthma. J Virol 2010: 84(15): 7418–7426.
3. Mackay IM. Human rhinoviruses: The cold wars resume. J Clin Virol 2008: 42: 297–320.
4. Sedlak RH, Nguyen T, Palileo I, Jerome KR, Kuypers J. Superiority of digital RT-PCR over real-time RT-PCR for quantitation of highly divergent human rhinoviruses. J Clin Microbiol 2017; 55(2): 442–449.
5. Andries K, Dewindt B, Snoeks J, Wouters L, Moereels H, Lewi PJ, Janssen PA. Two groups of rhinoviruses revealed by a panel of antiviral compounds present sequence divergence and differential pathogenicity. J Virol 1990: 64: 1117–1123.
6. Mackay IM, Arden KE, Nitsche A. Real-time PCR in virology. Nucleic Acids Res 2002: 30: 1292–1305.
7. Lu X, Holloway B, Dare RK, Kuypers J, Yagi S, Williams JV, Hall CB, Erdman DD. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J Clin Microbiol 2008: 46(2): 533–539.
8. Granados A, Luinstra K, Chong S, Goodall E, Bahn L, Mubareka S, Smieja M, Mahony J. Use of an improved quantitative polymerase chain reaction assay to determine differences in human rhinovirus viral loads in different populations. Diagn Microbiol Infect Dis 2012: 74: 384–387.
9. Tapparel C, Cordey S, Van Belle S, Turin L, Wai-Ming L, Regamey N, Meylan P, Mühlemann K, Gobbini F, Kaiser L. New molecular detection tools adapted to emerging rhinoviruses and enterviruses. J Clin Microbiol 2009: 47(6): 1742–1749.
10. Bochkov YA, Grindle K, Vang F, Evans MD, Gern JE. Improved molecular typing for rhinovirus species A, B, and C. J Clin Microbiol 2014: 52(7): 2461–2471.
11. Hoffman NG, Cook L, Atienza EE, Limaye AP, Jerome KR. Marked variability of BK virus load measurement using quantitative real-time PCR among commonly used assays. J Clin Microbiol 2008: 46(8): 2671–80.
12. Vynck M, Trypsteen W, Thas O, Vandekerckhove L, De Spiegelaere W. The future of the polymerase chain reaction in virology. Mol Diagn Ther 2016: 20: 437–447.
13. Huggett JF, Cowen S, Foy CA. Considerations for digital PCR as an accurate molecular diagnostic tool. Clin Chem 2015: 61: 79–88.
14. Sedlak RH, Jerome KR. Viral diagnostics in the era of digital polymerase chain reaction. Diagn Microbiol Infect Dis 2013: 75(1): 1–4.
The author
Jane Kuypers PhD
Department of Laboratory Medicine, University of Washington, Seattle, WA, USA
Vaccine and Infectious Disease Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA
E-mail: kuypers@uw.edu