Molecular diagnosis and sub-speciation of cutaneous leishmaniasis
Diagnosing cutaneous leishmaniasis histologically depends on the identification of the amastigotes, which is inconclusive and leads to cases of missed diagnosis or misdiagnosis. In this article, we describe a rapid diagnostic molecular method for Leishmania species identification and differentiation using DNA extracted from formalin-fixed paraffin-embedded (FFPE) skin tissue biopsies.
by L. Yehia and Dr I. Khalifeh
Clinical background
Cutaneous leishmaniasis is a chronic disease caused by Leishmania protozoan parasites that is on the increase in endemic and non-endemic regions because of environmental changes triggered by humans [1, 2]. It is most prevalent in the Middle East and North Africa. With changes in vector (sandfly), habitat and increased travel among populations, the incidence of leishmaniasis is showing a clear increase [3].
There are more than 20 strains of Leishmania that are pathogenic to humans [4], and these are partially responsible for its clinical diversity. The diagnosis of cutaneous leishmaniasis rests on the pathological identification of the amastigotes, which may be inconclusive [5]. This is dependent on the strain type, host response and the disease stage. Accurate microscopic diagnosis is essential to permit appropriate targeted therapy [6].
Clinically, cutaneous leishmaniasis may be asymptomatic and self-limiting. However, cases progressing to mutilating ulceration and disfiguring scarring have also been reported [7]. As the disease progresses, the number of amastigotes decreases to the point where none can be detected microscopically. The absence of amastigotes is a common problem encountered in up to 47% of cases [8]. In such instances, the diagnosis of cutaneous leishmaniasis must not be excluded [4].
Materials and methods
Skin biopsies embedded into FFPE tissue blocks were collected for 122 patients diagnosed clinically with cutaneous leishmaniasis. Cases included in the study were restricted to cutaneous lesions of patients who did not receive treatment prior to the biopsy. Cases with visceral or mucocutaneous involvement and with material insufficient for PCR or histopathological examination were excluded. Clinical information pertaining to the lesion was also collected including: number, duration, location and dermatologic appearance. In addition, the patient’s age, gender and country of residency were tabulated.
Cases were classified according to the modified Ridley’s parasitic index, a traditionally used pathological scoring system based on microscopic analysis of hematoxylin and eosin stained slides. DNA was then extracted from FFPE tissue blocks of each patient. Polymerase chain reaction (PCR) was performed using Leishmania-specific ribosomal internal transcribed spacer 1 (ITS1-PCR). Nested ITS1-PCR was performed on cases negative for conventional ITS1-PCR. ITS1-PCR amplicons were then digested with HaeIII for subsequent restriction fragment length polymorphism (RFLP) subspeciation.
Results
Of the 122 skin biopsies, microscopic evaluation of stained slides identified 54 cases (44.3%) labeled as histologically negative (with no unequivocal amastigotes detected). Of these negative cases, 9 (17%) were shave biopsies and 45 (83%) were punch biopsies.
DNA extracted from FFPE tissue blocks collected for all cases ranged from 4 to 1672 ng/μl (mean=213 ng/μl, SD=289 ng/μl). The oldest blocks were 19 years of age, whereas the newest were less than 1 year old. The quantity of the extracted DNA dating back to 1992 was 166 ng/μl (SD=128 ng/μl), whereas that for specimen from the year 2010 was 272 ng/μl (SD=161 ng/μl) indicating that a good quantity of DNA could be extracted from archival well-preserved FFPE tissues, even when they were old.
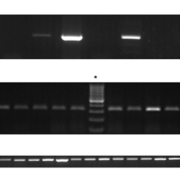
ITS1-PCR was performed on DNA extracted from all cases. Initially, and regardless of the histopathological analysis, 55 (45%) cases were positive and showed a band of between 300 and 350 base pairs indicative of Leishmania by agarose gel electrophoresis. The remaining 67 (55%) were negative (Fig. 1A, B). The negative cases were subjected to nested ITS1-PCR and 100% of these cases actually turned out to be positive for Leishmania (Fig. 1C).
Comparing the resultant ITS1-PCR bands to the DNA pattern of normal skin tissues, we identified 54 cases – that had been shown as negative by histopathology according to Ridley’s parasitic index – that amplified DNA with Leishmania-specific primers by conventional or nested ITS1-PCR, and that failed to show the normal skin profile seen in the negative controls tested. RFLP analysis identified L. tropica subspecies in all cases, identified by the presence of a 200 and 60 base pairs restriction fragments (Fig. 2) [9].
Clinical and diagnostic significance
Cutaneous leishmaniasis is a disease that is endemic in many regions of the world. With the ease of travel in the world, human and animal reservoirs of Leishmania parasites have been established in regions that previously were not known to harbour the sandfly vector because of habitat incompatibility. Thus, novel endemic areas have emerged in regions across the world. Therefore, a high index of suspicion becomes crucial for early diagnosis and control of leishmaniasis. With the advent of molecular diagnostic techniques and their high sensitivity and specificity, it has become easier to detect and control many infectious diseases, including leishmaniasis, as shown in this and other studies.
Traditionally, direct detection of parasites is performed by microscopic examination of clinical specimens or by cultivation, but either approach may be diagnostically problematic [1, 4, 10]. Cultures may take long periods, possibly weeks, for sufficient parasites to grow for species characterization. In addition, success in microscopic identification of amastigotes in stained preparations varies depending on the number of parasites present and/or the experience of the person examining the slide [11]. This is mainly due to the fact that all Leishmania species are morphologically similar and may present with a variable number of amastigotes. As the disease progresses, the number of amastigotes decreases to the point where none can be detected histopathologically.
Despite these drawbacks, microscopic identification and parasite cultivation are still the primary diagnostic tools used in most regions where leishmaniasis is endemic. However, it is stressed that accurate and rapid species identification is not possible using either technique. In the last decade, polymerase chain reaction (PCR) analysis has been successfully introduced and has been proven to be the most sensitive molecular tool for direct detection and parasite characterization of Leishmania species in clinical samples [1, 5, 12].
Accurate Leishmania species identification and subspeciation in clinical specimens is now possible by subjecting the extracted DNA to PCR, followed by enzymatic digestion to identify restriction fragments indicative of the subspecies. Such amplification using Leishmania-specific primers allows the indirect yet conclusive detection of the amastigotes, when present in a given clinical specimen. A highly sensitive method is valuable especially in chronic cases where the parasitic index is low and potentially undetectable by conventional microscopy.
Conclusion
This study successfully identified L. tropica in 54 skin biopsies from patients clinically suspected of having cutaneous leishmaniasis with negative biopsies. The importance of this result is manifested in the need for diagnostic tools that are sensitive, specific, rapid and capable of identifying all clinically significant Leishmania species from FFPE tissue blocks (Fig. 3).
Therefore, ITS1-PCR carried out on DNA extracted from FFPE tissue specimens, followed by HaeIII RFLP analysis, is a valuable method for the rapid and reliable diagnosis of cutaneous leishmaniasis. In chronic cases where the parasite load is low, or when insufficient tissue is available, nested ITS1-PCR can be performed to increase sensitivity. The advantages of this method are also highlighted with the possibility of using different biological specimens, and the ability to detect both Old World and New World leishmaniasis.
The work summarized here was first published as Yehia L. et al., 2012 [13].
References
1. Schonian G, et al. Diagn Microbiol Infect Dis 2003; 47: 349.
2. Goto H, Lindoso JA. Expert Rev Anti Infect Ther 2010; 8: 419.
3. Scarisbrick JJ, et al. Travel Med Infect Dis 2006; 4: 14.
4. Ameen M. Clin Exp Dermatol 2010; 35: 699.
5. Singh S, et al. Expert Rev Mol Diagn 2005; 5: 251.
6. Salman SM, et al. Clin Dermatol 1999; 17: 291.
7. David CV, Craft N. Dermatol Ther 2009; 22: 491.
8. Safaei A, et al. Dermatology 2002; 205: 18.
9. Kazemi-Rad E. Iran J Public Health 2008; 37: 54.
10. Farah FS, et al. Arch Dermatol 1971; 103: 467.
11. Bensoussan E, et al. J Clin Microbiol 2006; 44: 1435.
12. Schonian G, et al. Trends Parasitol 2008; 24: 135.
13. Yehia L, et al. J Cutan Pathol 2012; 39: 347–355.
The authors
Lamis Yehia, BSc
Biomedical Sciences Training Program, Case Western Reserve University in Cleveland, Ohio, USA
Ibrahim Khalifeh, MD
Department of Pathology and Laboratory Medicine, American University of Beirut, Beirut, Lebanon
E-mail: ik08@aub.edu.lb