Molecular subtypes of metastatic urothelial carcinoma and treatment options
Urothelial carcinoma is a common cancer and patients with metastatic urothelial cancer (mUC) have very poor outcomes. Many mUC patients do not have durable responses to systemic treatment and are exposed to toxic side effects that negatively impact the patient’s quality of life. Identifying novel biomarkers for therapy and selecting the best treatment option for individual mUC patients is therefore critical. Molecular characterization of urothelial carcinoma has revealed subtypes with different genomic, transcriptomic, and clinicopathological characteristics, which might guide therapeutic decisionmaking. Here, we summarize advances on molecular subtyping of urothelial carcinoma in both primary and metastatic disease, and highlight treatment opportunities for each subtype.
Background
Urothelial cancer (UC) of the urinary tract is the tenth most common malignant tumour worldwide affecting predominantly the bladder [1]. The main risk factor is attributed to smoking, although it has also been associated with parasitic infection and occupational exposure to chemicals. The incidence rate of bladder cancer is higher in men (9.5 per 100 000) than in women (2.4 per 100 000). However, women generally have more advanced disease at the time of diagnosis and response to treatment is less favourable [2]. Among men, it represents the sixth most common type of cancer and the ninth cause of cancer death.
Bladder cancer is a molecularly and clinically heterogeneous disease that is classified as either non-muscle-invasive bladder cancer (NMIBC) or muscle-invasive bladder cancer (MIBC). Approximately, 75% of bladder cancers are NMIBC, and these patients have excellent survival but suffer from high recurrence rates. The remaining 25% of patients have MIBC, which has a poor prognosis despite aggressive local and systemic treatment. Up to 50% of MIBC patients will develop metastasis [3], which is the primary cause of death. Therefore, it is essential to identify targets for therapy and to develop a more personalized treatment for mUC.
The rapid decrease in cost for next generation sequencing technologies in recent years has allowed researchers to genomically and transcriptomically characterize large cohorts of patients with (urothelial) cancer. This has significantly improved our knowledge on the molecular characteristics of cancer and has allowed us to explore new therapeutic options. Recently, the first study characterizing the genome and transcriptome of a large cohort of metastatic UC (mUC) was reported [4], which might contribute to the identification of therapeutic targets for this lethal disease.
Molecular landscape of urothelial carcinoma
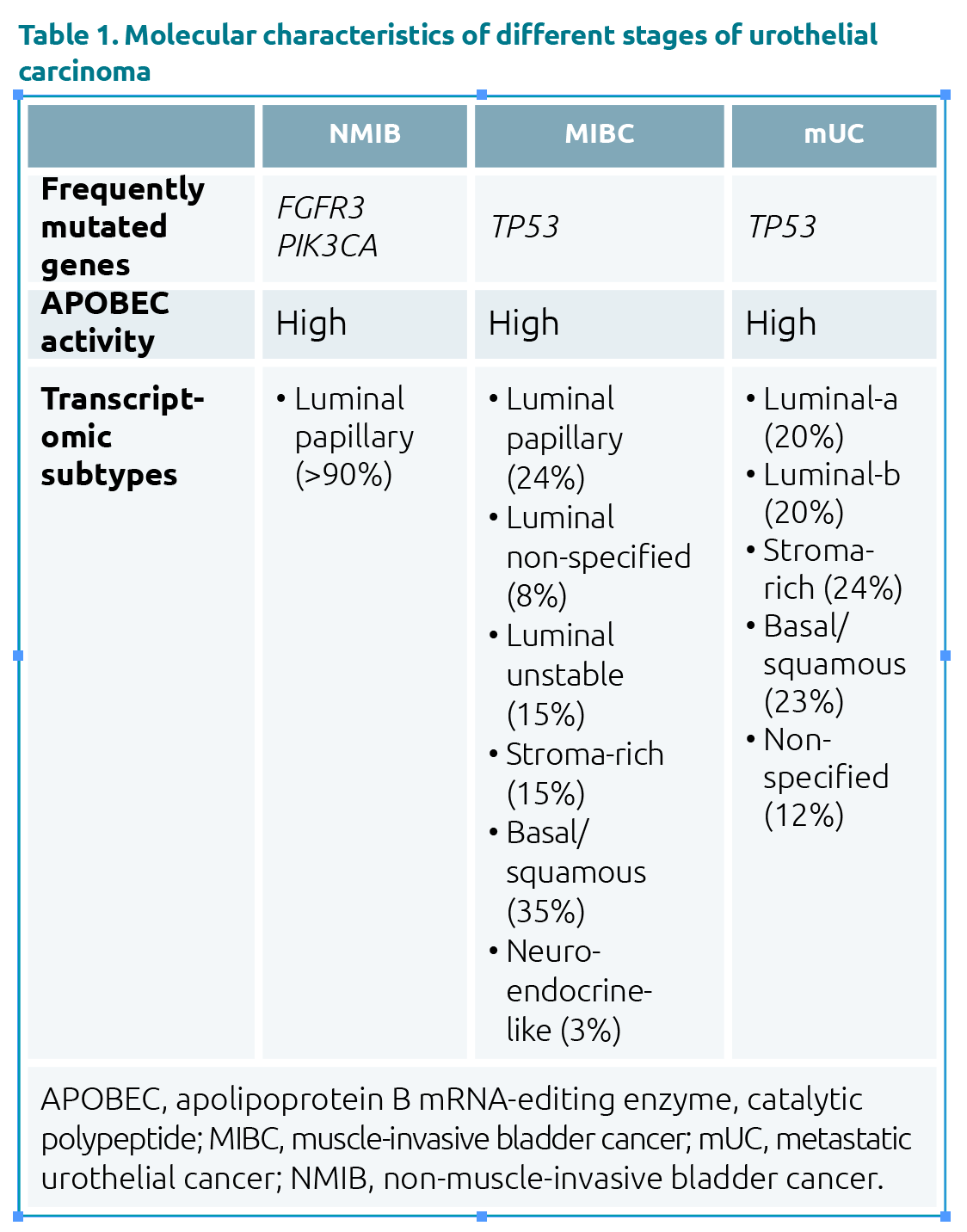
Comprehensive molecular profiling of UC has been extensively described for patients with NMIBC [5] and localized MIBC [6]. At the genomic level, NMIBC is characterized by frequent mutations in FGFR3 and PIK3CA genes, whereas mutations in TP53 are uncommon [7]. In MIBC, TP53 is the most commonly mutated gene (Table 1).
A recent study that investigated whole genomes and paired transcriptomes of a large cohort of patients with mUC, revealed genomic similarities between MIBC and mUC [4]. The set of driver genes identified in mUC and the high apolipoprotein B mRNA editing catalytic polypeptide-like (APOBEC) activity resembled the genomic landscape of MIBC.
At the transcriptomic level, NMIBC appears to be a more homogenous disease than MIBC or mUC (Table 1).
Genomic subtypes
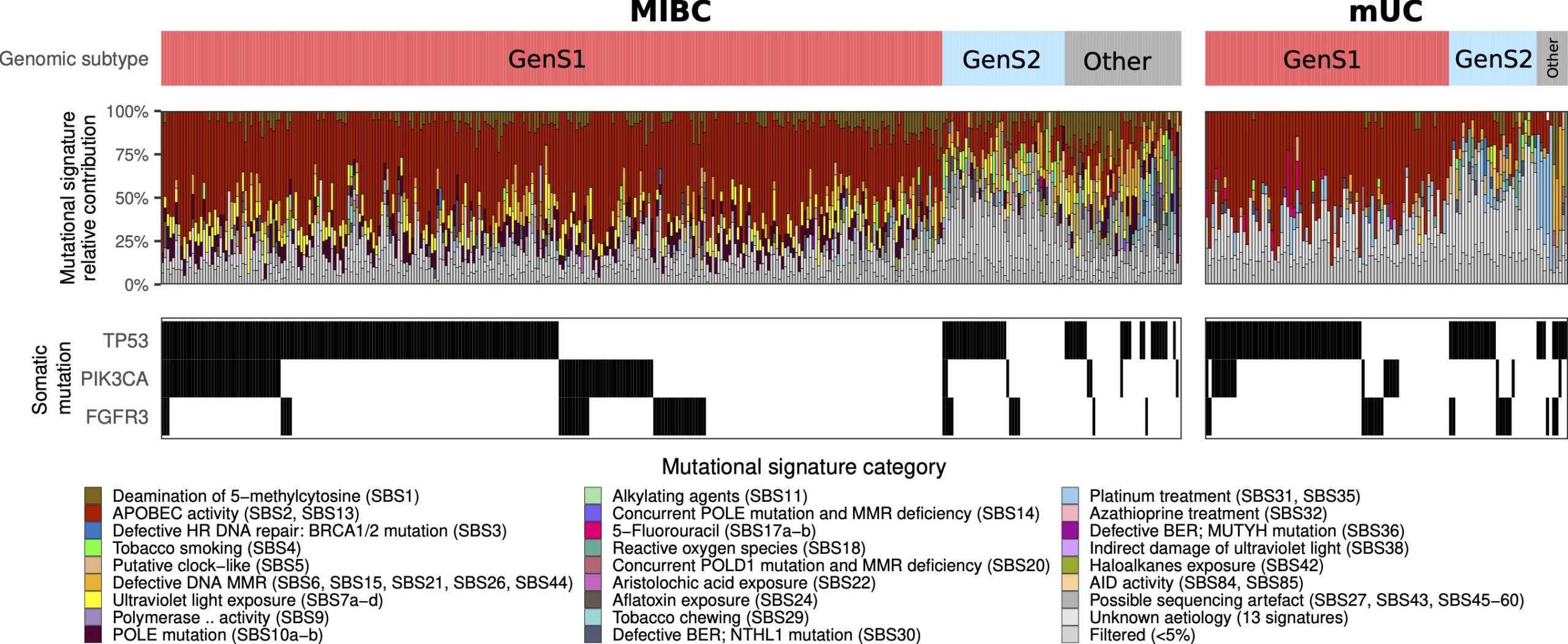
Cancer genomes accumulate mutations in a specific tri-nucleotide context that contain information regarding the mutagenic processes that a specific cancer has been exposed to. The patterns retrieved from these mutations are mutational signatures associated with specific etiologies. The clustering of mutational signatures revealed two major genomic subtypes (GenS) in primary MIBC and mUC (Fig. 1). The most prevalent subtype was GenS1, which was associated with APOBEC activity, and the second most prevalent subtype, GenS2, aggregated predominantly tumours characterized by mutational signatures associated with reactive oxygen species and are putatively clock like [4].
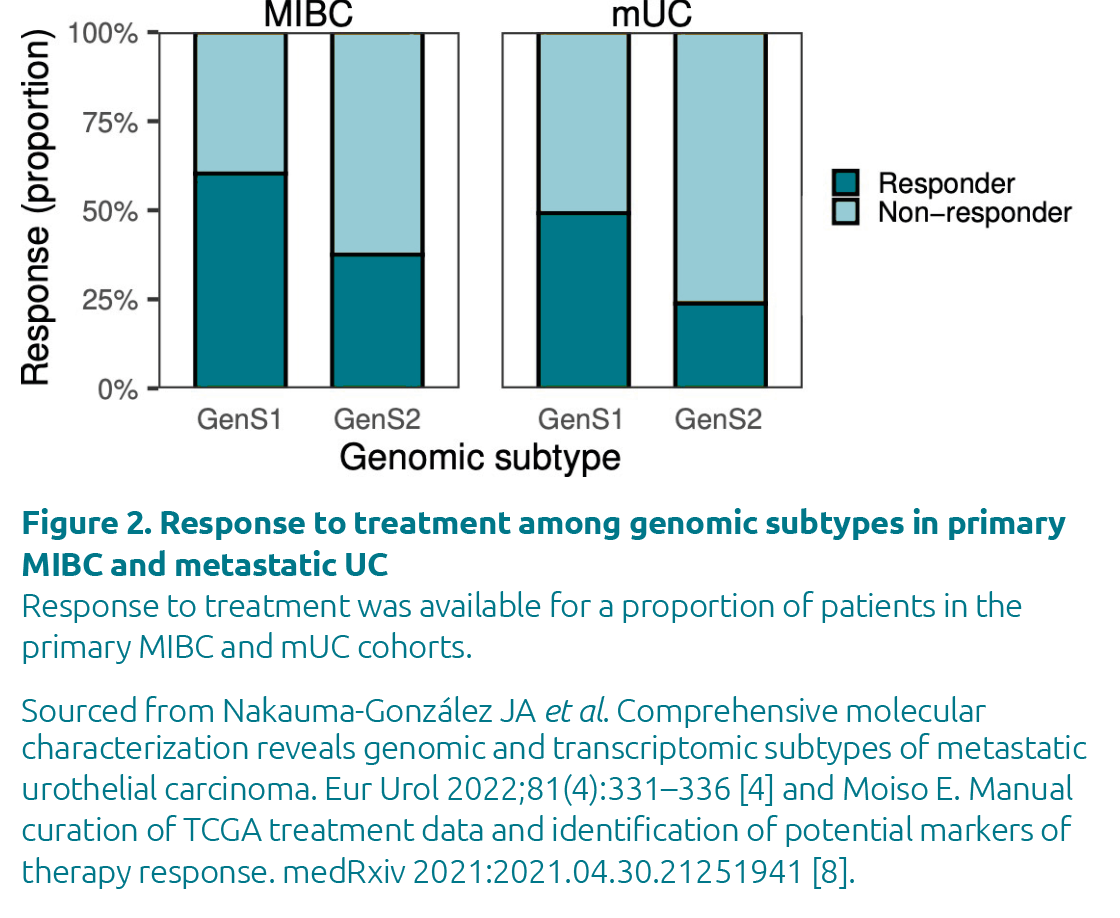
Previous studies showed that MIBC patients with APOBEC-driven tumours had a better outcome [6], which was also reflected in the GenS1 subtypes of both MIBC and mUC (Fig. 2).
Transcriptomic subtypes
An effort from the scientific community to establish a consensus transcriptomic classifier for MIBC recognized six subtypes in MIBC [9]. A tool was also developed to allow individual samples to be classified as one of the six consensus subtypes. This classification system was, however, exclusively developed for primary MIBC and it is not suitable for mUC subtyping [10]. It is important to stress that all samples from patients with primary MIBC are derived from the same organ, whereas biopsy samples from patients with mUC are derived from different metastatic sites. As a result, the background transcriptome may affect an accurate identification of the metastatic subtype. Owing to this limitation, a de novo transcriptomic subtyping system specific for mUC was developed that corrected for the origin of the metastatic biopsy [4].
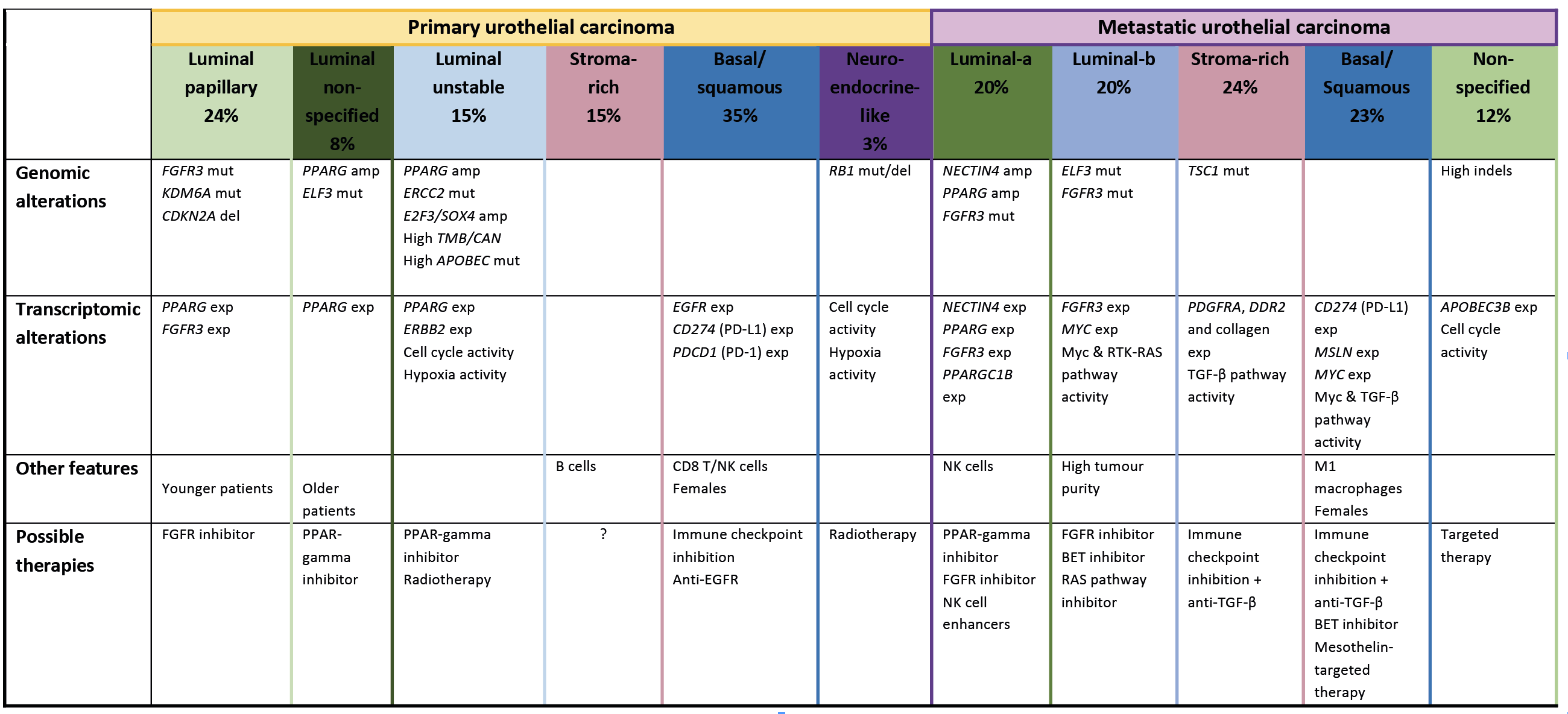
The new transcriptomic mUC subtyping system incorporated five transcriptomic subtypes. Each subtype had unique genomic and transcriptomic features. No neuroendrocrine-like (NE-like) subtype was identified in mUC even though it is known to be the most aggressive subtype of MIBC. The non-specified mUC subtype was enriched for patients previously treated with chemotherapy, and therefore it may represent a treatment resistance subtype. A similar subtype was not present in the consensus subtype system of primary MIBC as only chemotherapy naïve patients were considered. Although tumours with a luminal phenotype were present in MIBC and mUC, the specific luminal subtypes show distinct features in both diseases. The basal/squamous subtype had similar character-istics in both mUC and primary MIBC. This similarity also manifests in a poor prognosis for the consensus basal/squamous MIBC subtype and in a poor response to treatment in the basal/squamous mUC subtype [4,9].
Molecular subtypes and clinical implications
The genomic subtypes might be considered to select mUC patients for treatment. Patients with tumours of GenS1 could benefit from current available treatment options, whereas alternative treatments can be explored for other genomic subtypes. For example, the GenS2 subtype was enriched for tumours with homologous recombination deficiency and could potentially be a target for treatment with poly-ADP ribose polymerase inhibitors and/or double-stranded DNA break-inducing chemotherapy [4].
The development of the transcriptomic subtype systems revealed potential therapeutic strategies per subtype (Fig. 3). In both mUC and MIBC, the basal/squamous subtype could be sensitive to immune checkpoint inhibition as immune checkpoint markers were highly expressed. For other transcriptomic subtypes, therapeutic options might vary for mUC and for MIBC.
Future perspectives
Molecular subtyping of urothelial carcinoma revealed extensive heterogeneity of this disease in the metastatic setting. Specifically, the transcriptomic subtypes showed distinct characteristics that might guide targeted therapy. These subtypes, however, have to be validated in other independent cohorts with the aim of developing a consensus classification system for mUC. It is also essential to clarify if selecting patients for treatment based on molecular subtypes results in a better outcome for patients with mUC. A recent study, the COXEN trial, that aimed to prospectively validate a predictive transcriptomic-based biomarker in 167 patients with primary MIBC was conducted [11]. However, the trial did not meet its primary endpoint, which was to predict pathological response to neo-adjuvant chemotherapy. This study demonstrated the challenges ahead for the clinical implementation of transcriptomic subtyping platforms. These challenges will only be bigger in the metastatic setting because the majority of mUC patients received several lines of (systemic) treatment before diagnosis with metastatic disease. It is unclear how treatment could influence the transcriptomic profile of a tumour, although evidence suggests that therapeutic pressure may induce a dramatic change in the identity of a tumour through a phenotype-switching event [12]. Thus, treatment history would be a key variable to consider in patients with mUC to further refine the transcriptomic subtypes of mUC.
Thus far, the molecular subtype classification systems for urothelial carcinoma have prognostic value and offer opportunities for patient stratification. The COXEN trial is the only prospective study that has tested the predictive value of transcriptomic markers in primary MIBC and several ongoing clinical trials will eventually determine the clinical benefit of transcriptomic subtypes. Tools to identify transcriptomic subtypes are now (commercially) available [4,9,13], which could accelerate the implementation of molecular subtyping in the clinic if the outcome of prospective clinical trials is positive.
Figure 1. Genomic subtypes of primary MIBC and mUC
The bladder cancer cohort of The Cancer Genome Atlas (TCGA) was used to identify genomic subtypes of MIBC. Genomic subtypes, mutational
signatures and somatic mutations (single nucleotide variants and small insertions/deletions) in TP53, PIK3CA and FGFR3 are displayed.
Figure 3. Characteristics and treatment options for transcriptomic subtypes of urothelial carcinoma
A summary of relevant molecular and clinical characteristics, and therapeutic options per transcriptomic subtypes in primary MIBC and mUC.
Notes: Amp, amplification; BET, bromodomain and extra-terminal domain (protein); Del, deletion; EGFR, epidermal growth factor receptor; Exp, expression; FGFR, fibroblast growth factor; Indel, insertion–deletion mutations; Mut, mutation; Myc, Myc proto-oncogene protein; NK, natural killer; PD-1, programmed cell death protein 1; PD-L1, programmed cell death 1 ligand 1; PPARγ, peroxisome proliferator-activated receptor gamma; RAS, small GTPase proteins; TGF-β, transforming growth factor beta.
Sourced from Nakauma-González JA et al. Comprehensive molecular characterization reveals genomic and transcriptomic subtypes of metastatic urothelial carcinoma. Eur Urol 2022;81(4):331–336 [4] and Kamoun A et al. A consensus molecular classification of muscle-invasive bladder cancer. Eur Urol 2020;77(4):420–433 [9].
The authors
J. Alberto Nakauma-González1,2,3 PhD and Joost L. Boormans*2 MD, PhD
1 Cancer Computational Biology Center, Erasmus MC Cancer Institute, University Medical Center Rotterdam
2 Department of Urology, Erasmus MC Cancer Institute, University Medical Center Rotterdam
3 Department of Medical Oncology, Erasmus MC Cancer Institute, University Medical Center Rotterdam
*Corresponding author
E-mail: j.boormans@erasmusmc.nl
References
1. Sung H, Ferlay J, Siegel RL et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–249 (https://acsjournals.onlinelibrary.wiley.com/doi/10.3322/caac.21660).
2. Dobruch J, Daneshmand S, Fisch M et al. Gender and bladder cancer: a collaborative review of etiology, biology, and outcomes. Eur Urol 2016;69:300–310.
3. Stein JP, Lieskovsky G, Cote R et al. Radical cystectomy in the treatment of invasive bladder cancer: long-term results in 1,054 patients. J Clin Oncol 2001;19:666–675.
4. Nakauma-González JA, Rijnders M, van Riet J et al. Comprehensive molecular characterization reveals genomic and transcriptomic subtypes of metastatic urothelial carcinoma. Eur Urol 2022;81(4):331–336 (https://doi.org/10.1016/j.eururo.2022.01.026).
5. Lindskrog SV, Prip F, Lamy P et al. An integrated multi-omics analysis identifies prognostic molecular subtypes of non-muscle-invasive bladder cancer. Nat Commun 2021;12(1):2301 (https://www.nature.com/articles/s41467-021-22465-w).
6. Robertson AG, Kim J, Al-Ahmadie H et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell 2017;171:540–556.e25 (https://doi.org/10.1016/j.cell.2017.09.007).
7. Knowles MA, Hurst CD. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat Rev Cancer 2015;15(1):25–41.
8. Moiso E. Manual curation of TCGA treatment data and identification of potential markers of therapy response. medRxiv 2021:2021.04.30.21251941 (https://www.medrxiv.org/content/10.1101/2021.04.30.21251941v1.full-text).
9. Kamoun A, de Reyniès A, Allory Y et al. A consensus molecular classification of muscle-invasive bladder cancer. Eur Urol 2020;77(4):420–433
(https://doi.org/10.1016/j.eururo.2019.09.006).
10. Williams SB, Black PC, Dyrskjøt L et al. Re: Aurélie Kamoun, Aurélien de Reyniès, Yves Allory, et al. A consensus molecular classification of muscle-invasive bladder cancer. Eur Urol 2020;77:420–433: A statement from the International Bladder Cancer Network. Eur Urol 2020;77(4):e105–e106.
11. Flaig TW, Tangen CM, Daneshmand S et al. A randomized phase ii study of coexpression extrapolation (COXEN) with neoadjuvant chemotherapy for bladder cancer (SWOG S1314; NCT02177695). Clinical Cancer Research 2021;27(9):2435–2441.
12. Marine JC, Dawson SJ, Dawson MA. Non-genetic mechanisms of therapeutic resistance in cancer. Nat Rev Cancer 2020;20(12):743–756.
13. GenomeDx launches decipher bladder cancer classifier. UroToday 2017, 10 May (https://www.urotoday.com/recent-abstracts/urologic-oncology/bladder-cancer/95552-genomedx-launches-decipher-bladder-cancer-classifier.html)