Mumps virus sequencing methods for high-resolution outbreak analysis
Despite the enormous success of mumps vaccination campaigns around the world, there has recently been a significant increase in mumps cases and outbreaks in vaccinated populations. As a result, tracking the molecular epidemiology of mumps outbreaks has increased in importance. Here we examine the challenges and limitations associated with traditional molecular genotyping tools and compare the different mumps whole-genome sequencing techniques for molecular surveillance and outbreak tracing.
Introduction
Many readers of this article are likely to have a senior member of their family that was diagnosed with mumps, usually in their childhood. If you ask them what it was like, they will probably describe a painful swelling on one or both sides of their face. This swelling, known as parotitis due to inflammation of salivary glands such as the parotid gland, is often what we imagine when we think of mumps. In addition, they may recall having a fever, a very bad sore throat and malaise. Hopefully they did not suffer any of the rarer, more serious complications that can occur such as sterility, hearing loss and encephalitis [1].
Mumps is a highly contagious infectious disease caused by a singlestranded, negative-sense, enveloped RNA virus of the genus Rubulavirus in the family Paramxyoviridae [2]. Mumps was an incredibly common disease in the pre-vaccine era with a global incidence greater than 100 cases per 100 000 population annually [3].
However, following the introduction of mumps vaccination programs these numbers began to plummet, resulting in an incidence of only 0.1 cases per 100 000 population by 2001 [4]. This achievement caused a great deal of excitement in the public health community, raising hopes for the implementation of mumps eradication programs.
Unfortunately, mumps has begun to re-emerge increasing, both in the number of mumps cases and the number of outbreaks [5–8]. Of additional concern, this re-emergence is occurring globally and in fully vaccinated individuals, often associated with close-quarter settings such as schools and universities [9]. The exact cause for this increase remains controversial but both waning immunity and vaccine evasion have been suggested [10]. What is certain, is that these outbreaks have placed a significant burden on public health infrastructure by increasing the volume of laboratory testing and the number and scope of epidemiological investigations.
Molecular diagnostics and surveillance
Laboratory testing plays an important role in mumps diagnosis, as patients can be asymptomatic or develop other symptoms without presenting with parotitis. Conversely, the presence of parotitis does not guarantee mumps infection, as parotitis can be caused by other viruses including influenza, parainfluenza, and Epstein-Barr [11].
Diagnostic testing commonly includes serology for mumps IgM antibodies and real-time RT-PCR for mumps virus nucleic acid detection. Real-time RT-PCR is especially important in vaccinated populations because individuals infected with the mumps virus can sometimes fail to produce detectable levels of IgM antibodies [12].
Molecular surveillance for mumps is performed by countries around the world using traditional sequencing methods on the viral nucleic acid obtained from positive samples. This typically involves sequence analysis of a short stretch of 316 nucleotides in the highly variable mumps small-hydrophobic (SH) gene. The WHO used this method in part to devise an alphabetic mumps genotyping scheme consisting of twelve genotypes designated A–N and excluding E and M [13].
The standardization and implementation of this genotyping scheme was very effective for describing global mumps diversity and monitoring mumps transmission pathways for many years [14]. However, the success of mumps vaccination programs has caused a bottleneck in the genetic diversity of the remaining mumps viruses. Most mumps viruses detected now are genotype G and even between distinct outbreaks, few (0–5) single nucleotide polymorphisms in the SH gene are identified with which to differentiate the genotype G strains [15]. Unfortunately, the decrease in genetic heterogeneity has relegated the utility of these traditional sequencing methods to monitoring global genotype dynamics rather than outbreak analysis within countries.
Efforts have been made to combine sequences from several mumps genomic regions including the SH, HN, and F genes to provide meaningful epidemiological information [16]. However, this requires sequencing and concatenating multiple gene targets with conventional sequencing methods that are often laborious and time-consuming. Given the significant homogeneity of the currently circulating genotype G strains, these combined sequencing approaches cannot match the molecular resolution and efficiency of whole-genome sequencing (WGS).
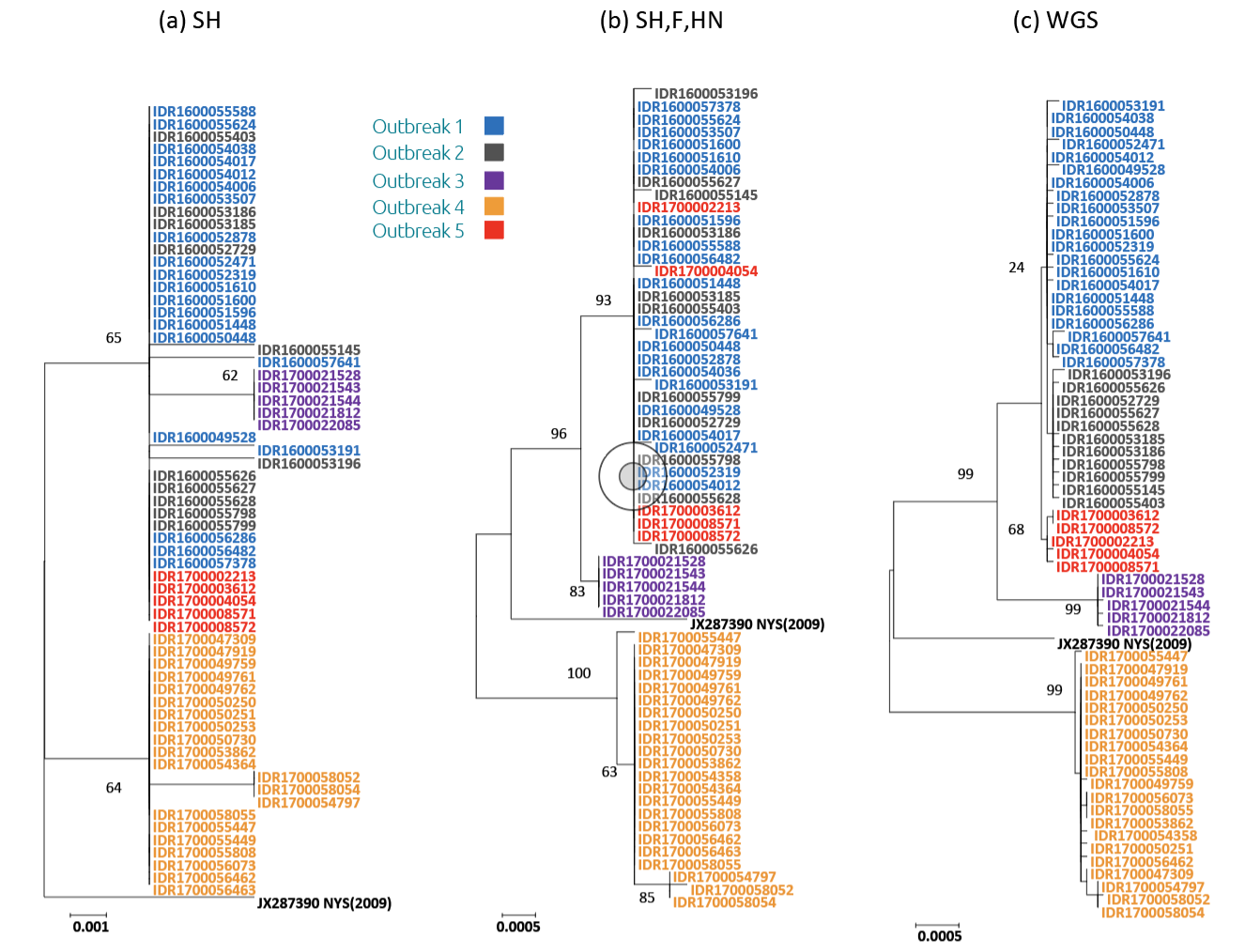
We recently demonstrated these resolution differences in a study that compared the phylogenetic trees generated with sequence data from the SH gene alone (Fig. 1a), concatenated sequences from the SH, HN, and F genes (Fig. 1b), and mumps whole-genome sequences (Fig. 1c) [17]. We used a custom sequencing panel to generate the whole-genome sequences from specimens selected from several outbreaks across New York State, USA. Importantly, while the three gene approach was able to accurately differentiate most of the outbreaks, only the WGS approach was able to delineate more closely related outbreaks. In addition to providing high-resolution analysis for outbreaks, the data generated from mumps WGS has also been shown to facilitate the estimation of transmission links between individual cases [18, 19].
Mumps WGS assays
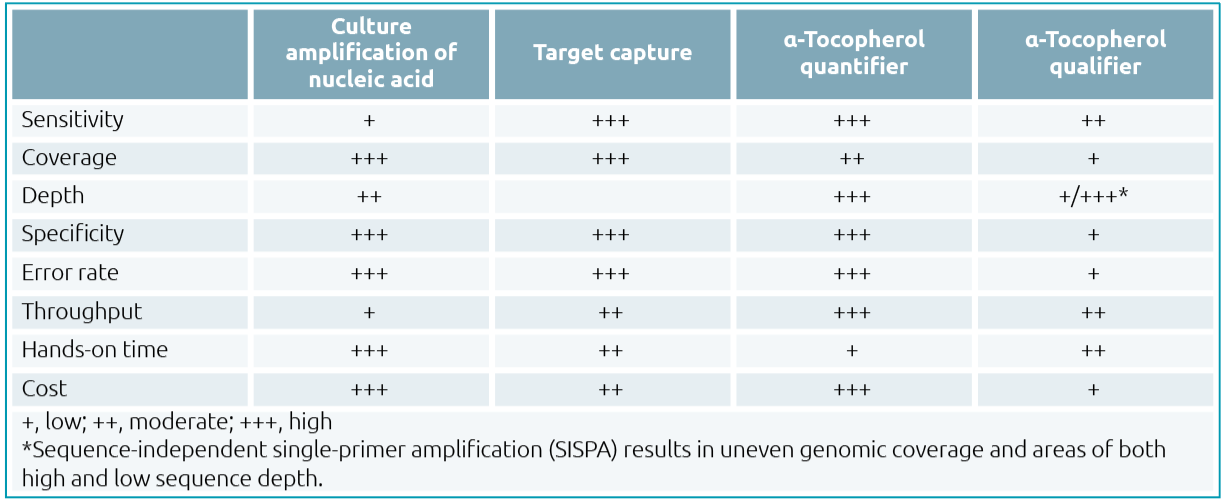
Scientists interested in establishing a mumps WGS assay in their laboratory may feel overwhelmed by the variety of methods that are currently available. All come with inherent pros and cons, some of which are discussed here and summarized in Table 1. Although sequencing platforms, sequence analysis software and bioinformatics pipelines are all integral components of the process, this brief review will focus on methods commonly used for generating the mumps nucleic acid needed for sequencing.
Culture amplification
First, large quantities of mumps nucleic acid can be produced by initially culturing the virus from positive patient specimens in replication competent cell lines [20]. After the virus has grown, mumps nucleic acid can be harvested and extracted in large quantities, then carried through the additional downstream steps in the sequencing process. Although this non-molecular method has the advantage of not requiring initial viral nucleic acid enrichment by PCR amplification, it does require technical knowledge, experience and the necessary laboratory resources to safely generate cultured viral isolates. In addition, although this method circumvents PCR-amplification-induced errors, genomic mutations have been shown to occur, with approximately two nucleotide changes per wholegenome having been found during viral replication [21].
Target capture
The second method is often referred to as target capture or ‘catch’ because the mumps virus nucleic acid in the primary specimen is hybridized to labelled (typically biotinylated) mumps-specific oligonucleotide probes, or ‘bait’ [22]. The resulting mumps RNA isolated at the end of the procedure is both purified and enriched. Since this technique does not require viral culture isolation or nucleic acid amplification by PCR, the whole-genome sequences produced are free from amplification-induced errors and are considered the most ‘unbiased’. However, this procedure requires technical molecular knowledge concerning ‘bait’ design, although virus-specific ‘bait’ probes are becoming increasingly commercially available [23]. In addition, the protocols themselves are laborious and time-consuming as several sample treatment steps are typically required before hybridization to remove unwanted proteins, lipids, nucleic acid and other interfering substances that minimize viral-specific RNA yield.
Amplicon tiling
The third method, known as amplicon tiling or ‘jackhammer’, uses specific primer sets to generate overlapping amplicons of the mumps genome. This approach commonly has enhanced sensitivity since the mumps nucleic acid is amplified by PCR, which ultimately results in successful sequencing even when applied directly to specimens with low viral loads. These assays can also be highly automated as preamplification processing is not necessary, significantly reducing the hands-on time. Post-sequencing processing and data analysis can also be streamlined by the instrument software, thereby decreasing the amount of manual analysis required [24]. However, this method can be expensive, with costs proportional to the amount of automation, disposables, and reagents used. Also, because tiling methods are PCR-based, they require prior knowledge of the target sequence. Furthermore, the number of primer sets required can become substantial as the genetic heterogeneity of the target organism increases. The requirement for PCR will also increase the potential for more amplification-induced sequencing artifacts, although these errors continue to be minimized as amplification, sequencing and analysis technologies improve. Another limitation is that primer binding on the ends of the genomes inherently results in a lack of sequence coverage at the 5′ and 3′ termini of the genome. This can be overcome by using a genetically independent or ‘agnostic’ approach known as sequence-independent single-primer amplification, or ‘SISPA’ [25].
SISPA
The fourth method, SISPA, uses non-specific random primers to amplify the RNA present in the sample, so that prior knowledge of viral target sequences is not required. This has made SISPA especially useful for identifying new viruses, but the technology has drawbacks when attempting to amplify complete genomes. For instance, the method has been shown to produce spotty coverage across some genomes, and to increase transition-event-induced errors [26]. This can result in highly variable sequencing depth across some genomes. As with the ‘bait’ method described above, SISPA can also require a significant amount of sample treatment and preparation to remove unwanted and interfering substances. As a result, the method is rarely used for routine mumps sequencing but may be useful in situations where the genetic diversity of the mumps samples is particularly high [27].
Summary
In conclusion, the emergence of mumps viruses with limited genetic heterogeneity in vaccinated populations has resulted in a need to generate viral whole-genome sequences rapidly and efficiently. Applications for this sequence data include molecular epidemiological investigations, surveillance of emerging variants with vaccine evasion potential, and the design and modification of viral detection assays. Although there are several sequencing approaches to choose from, all of them have advantages and disadvantages, therefore scientists interested in establishing a mumps WGS assay in their laboratory should consider the option that best meets their resources, molecular expertise, laboratory capacity, anticipated testing load and infrastructure.
Acknowledgments
We thank the Applied Genomics Technology Core at the Wadsworth Center for their help in performing dideoxy sequencing. We also thank the Bureau of Immunization at the New York State Department of Health for providing epidemiological information.
This work was supported in part by Cooperative Agreement Number NU60OE000104, funded by the Centers for Disease Control and Prevention through the Association of Public Health Laboratories. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the Centers for Disease Control and Prevention, the Department of Health and Human Services, or the Association of Public Health Laboratories. PB, HC, DL and TY declare no competing interests. KSG receives research support from ThermoFisher for the evaluation of new assays for the detection and characterization of viruses. She also has a royalty-generating collaborative agreement with Zeptometrix.
Figure 1. Phylogenetic comparison of sequences from 63 mumps viruses from several outbreaks that occurred in New York State from 2016 to 2017
Samples from individual outbreaks are identified by specific colors, as indicated by the outbreak key. Sequence alignments were generated using the SH gene of the mumps genome (a), concatenated sequences of the SH, F, and HN genes (b), or the mumps whole-genome sequence (c). The evolutionary history was inferred by using the maximum likelihood method and the Tamura-Nei model. The phylogeny was tested using 1000 bootstrap replicates; values of >75 are indicated on the tree. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Evolutionary analyses were conducted in MEGA X.
Note: This figure was reprinted from Bryant P, Caldwell H, Lamson DM, Yildirim T, St George K. 2022. Streamlined whole-genome sequencing of mumps virus for high-resolution outbreak analysis. J Clin Microbiol 60:e0084121 [17] with permission from the Journal of Clinical Microbiology) and permission to re-use the figure was granted by the Journals Department of the Journal of Clinical Microbiology.
Table 1. Advantages and disadvantage of mumps nucleic acid enrichment methods for whole-genome sequencing
The authors
Patrick W Bryant*1, Haley S Caldwell1,2,3, Daryl M Lamson1, and Kirsten St. George1,2
1Laboratory of Viral Diseases, Wadsworth Center, New York State Department of Health, Albany, New York, 12208 USA
2Department of Biomedical Science, University at Albany, SUNY, Albany, New York, USA
3The Arbovirus Laboratory, Wadsworth Center, New York State Department of Health, Slingerlands, New York, USA
*Corresponding author
E-mail: patrick.bryant@health.ny.gov
References
- Brook I. Diagnosis and management of parotitis. Arch Otolaryngol Head Neck Surg 1992;118:469–471.
- Rima BK, Wishaupt RG, Welsh MJ, Earle JA. The evolution of morbilliviruses: a comparison of nucleocapsid gene sequences including a porpoise morbillivirus
Vet Microbiol 1995;44:127–134. - Galazka AM, Robertson SE, Kraigher A. Mumps and mumps vaccine: a global review. Bull World Health Organ 1999;77:3–14 (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2557572/pdf/10063655.pdf).
- McNabb SJ, Jajosky RA, Hall-Baker PA et al. Summary of notifiable diseases — United States, 2005. MMWR Morb Mortal Wkly Rep 2007;54:1–92 (https://www.cdc.gov/mmwr/preview/mmwrhtml/mm5453a1.htm).
- Dayan GH, Quinlisk MP, Parker AA et al. Recent resurgence of mumps in the United States. N Engl J Med 2008;358:1580–1589 (https://www.nejm.org/doi/10.1056/NEJMoa0706589?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%20%200www.ncbi.nlm.nih.gov).
- Bag SK, Dey A, Wang H, Beard F. Australian vaccine preventable disease epidemiological review series: mumps 2008–2012. Commun Dis Intell Q Rep 2015;39:E10–18 (https://www1.health.gov.au/internet/main/publishing.nsf/Content/cda-cdi3901b.htm).
- Peltola H, Kulkarni PS, Kapre SV et al. Mumps outbreaks in Canada and the United States: time for new thinking on mumps vaccines. Clin Infect Dis 2007;45:459–466 (https://academic.oup.com/cid/article/45/4/459/425811?login=false).
- Sane J, Gouma S, Koopmans M et al. Epidemic of mumps among vaccinated persons, The Netherlands, 2009–2012. Emerg Infect Dis 2014;20:643–648 (https://wwwnc.cdc.gov/eid/article/20/4/13-1681_article).
- Connell AR, Connell J, Leahy TR, Hassan J. Mumps outbreaks in vaccinated populations – Is it time to re-assess the clinical efficacy of vaccines? Front Immunol 2020;11:2089 (https://www.frontiersin.org/articles/10.3389/fimmu.2020.02089/full).
- Dayan GH, Rubin S. Mumps outbreaks in vaccinated populations: are available mumps vaccines effective enough to prevent outbreaks? Clin Infect Dis 2008;47:1458–1467 (https://www.frontiersin.org/articles/10.3389/fimmu.2020.02089/full).
- Elbadawi LI, Talley P, Rolfes MA et al. Non-mumps viral parotitis during the 2014–2015 influenza season in the United States. Clin Infect Dis 2018;67:493–501 (https://academic.oup.com/cid/article/67/4/493/4957004?login=false).
- Jin L, Vyse A, Brown DW. The role of RT-PCR assay of oral fluid for diagnosis and surveillance of measles, mumps, and rubella. Bull World Health Organ 2002;80:76–77 (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2567626/pdf/11884979.pdf).
- Jin L, Rima B, Brown D et al. Proposal for genetic characterisation of wild-type mumps strains: preliminary standardisation of the nomenclature. Arch Virol 2005;150:1903–1909.
- Jin L, Orvell C, Myers R et al. Genomic diversity of mumps virus and global distribution of the 12 genotypes. Rev Med Virol 2015;25:85–101.
- McNall RJ, Wharton AK, Anderson R et al. Genetic characterization of mumps viruses associated with the resurgence of mumps in the United States: 2015–2017. Virus Res 2020;281:197935.
- Gouma S, Cremer J, Parkkali S et al. Mumps virus F gene and HN gene sequencing as a molecular tool to study mumps virus transmission. Infect Genet Evol 2016;45:145–150.
- Bryant P, Caldwell H, Lamson DM et al. Streamlined whole-genome sequencing of mumps virus for high-resolution outbreak analysis. J Clin Microbiol 2022;60:e0084121.
- Wohl S, Metsky HC, Schaffner SF et al. Combining genomics and epidemiology to track mumps virus transmission in the United States. PLoS Biol 2020;18:e3000611 (https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3000611).
- Stapleton PJ, Eshaghi A, Seo CY et al. Evaluating the use of whole genome sequencing for the investigation of a large mumps outbreak in Ontario, Canada. Sci Rep 2019;9:12615 (https://www.nature.com/articles/s41598-019-47740-1).
- Dilcher M, Barratt K, Douglas J et al. Monitoring viral genetic variation as a tool to improve molecular diagnostics for mumps virus. J Clin Microbiol 2018;56(10):e00405–18 (https://journals.asm.org/doi/10.1128/JCM.00405-18?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%20%200pubmed).
- Bodewes R, Reijnen L, Kerkhof J et al. Molecular epidemiology of mumps viruses in the Netherlands, 2017–2019. PLoS One 2020;15:e0233143 (https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0233143).
- Metsky HC, Siddle KJ, Gladden-Young A et al. Capturing sequence diversity in metagenomes with comprehensive and scalable probe design. Nat Biotechnol 2019;37:160–168 (https://www.nature.com/articles/s41587-018-0006-x).
- Frost JR, Schulz H, McLachlan E et al. An enrichment method for capturing mumps virus whole genome sequences directly from clinical specimens. J Virol Methods 2021;294:114176 (https://www.sciencedirect.com/science/article/pii/S0166093421001154?via%3Dihub).
- Plitnick J, Griesemer S, Lasek-Nesselquist E et al. Whole-genome sequencing of SARS-CoV-2: assessment of the ion torrent AmpliSeq Panel and comparison with the Illumina MiSeq ARTIC Protocol. J Clin Microbiol 2021;59:e0064921 (https://journals.asm.org/doi/full/10.1128/JCM.00649-21?rfr_dat=cr_pub++0pubmed&url_ver=Z39.88-2003&rfr_id=ori%3Arid%3Acrossref.org).
- Reyes GR, Kim JP. Sequence-independent, single-primer amplification (SISPA) of complex DNA populations. Mol Cell Probes 1991;5:473–481.
- Parras-Moltó M, Rodríguez-Galet A, Suárez-Rodríguez P, López-Bueno A. Evaluation of bias induced by viral enrichment and random amplification protocols in metagenomic surveys of saliva DNA viruses. Microbiome 2018;6:119 (https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-018-0507-3).
- Magana LC, Espinosa A, Marine RL et al. Complete genome sequences of mumps and measles virus isolates from three states in the United States. Genome Announc 2017
;5(33):e00748–17 (https://journals.asm.org/doi/full/10.1128/genomeA.00748-17?rfr_dat=cr_pub++0pubmed&url_ver=Z39.88-2003&rfr_id=ori%3Arid%3Acrossref.org).