Next-generation sequencing as a diagnostic tool in respiratory infections
Background
The term ‘acute respiratory tract infection’ (ARTI) encompasses a spectrum of conditions ranging from the common cold to pneumonia. Multiple organisms may infect the human respiratory tract, such as bacteria and fungi, but the majority of episodes are thought to be viral in origin [1]. The diagnosis of ARTI is made clinically but the lack of pathognomonic features means etiology cannot be determined clinically. Identifying causative organisms is challenging owing to the number of possibilities but is important for many reasons. In severe cases identification of an organism will guide therapeutics, although specific treatments for viral ARTI are generally limited to influenza. At the other end of the clinical spectrum identifying a viral cause in mild cases allows the physician to defer treatment with antibiotics and reassure the patient. This is an essential component of antimicrobial stewardship as high rates of antibiotic use are associated with circulating antimicrobial resistance [2]. The majority of antibiotic prescriptions issued in the UK are for respiratory tract infections [3] yet antibiotic use puts patients at risk of adverse drug reactions and in many cases will not lessen the duration of symptoms [4].
Respiratory infection diagnostics
Until recently viral diagnostics relied on cell culture or animal/egg inoculation. These time-consuming and laborious methods provided only a retrospective diagnosis and were, therefore, of little use in the management of acute infections. Nucleic acid amplification tests (NAAT), directly detecting the RNA or DNA of pathogens, have largely superseded these.
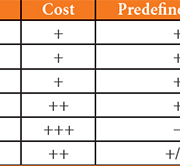
Many methods of RNA/DNA detection are available (Table 1). The most widely used is polymerase chain reaction (PCR). This has the benefit of rapid turnaround times and high levels of sensitivity and specificity in comparison to cell culture [5]. A pair of primers is required for each target but it is possible to use multiple primer sets within a single reaction (up to four) without compromising test sensitivity over the monoplex assay. This chemistry is now available as closed systems providing rapid results as a near-patient test (GeneXpert by Cepheid, Cobas by Roche).
Other examples of molecular NAATs which are available but not commonplace would be loop-mediated isothermal amplification (LAMP) and microarrays. LAMP detects nucleic acids but does not rely on thermocycling. It does, however, require multiple primers for each target (usually six) and as a consequence the sensitivity is more likely to be affected by genome mutations than standard PCR. This also makes multiplexing multiple targets within a single reaction more complicated.
Microarrays (also known as DNA chips or biochips) use a collection of oligonucleotide probes, about 70 bases in length, immobilized on a solid surface. The probes are complementary portions of DNA or RNA designed to match conserved regions of a genome; thus, if present, the target will bind to the corresponding probe which can then be quantified. Multiple probes may be attached to a single surface, screening for a large number of pathogens in a single reaction. As probes are targeted against conserved regions of the pathogen genome they may also detect related but novel pathogens.
To be used as a comprehensive diagnostic test numerous targets must be included to cover the likely pathogens. In the case of respiratory infections commercial assays are available with around 33 targets over 8 reactions [6]. Despite this approach a viral pathogen is detected in only a minority of specimens [7] and it remains the case that a pathogen will only be detected if actively sought.
Several significant respiratory viruses have been identified in recent years; those which may have circulated for many years, such as the human metapneumovirus, or emerging pathogens, such as SARS and, more recently, MERS. Whereas these are related to other known pathogens they are genetically distinct and, therefore, would evade detection with molecular methods.
What is next-generation sequencing?
The term ‘next-generation sequencing’ (NGS) refers to the practice of sequencing millions of DNA fragments in parallel. Numerous platforms are available to carry this out and the exact chemistry varies greatly between each. In practice, either all genetic material within a sample can be sequenced – metagenomics; or, hybrid capture allows a more focused approach to an area or genome of interest, this is termed ‘target enrichment’.
Advantages of NGS
Applying metagenomic NGS to clinical samples would allow an untargeted approach to identify all the genetic material contained within. This method has demonstrated potential for use in a diagnostic setting [8, 9].
The lack of pathogen targeting means that multiple pathogens can be detected without selection (Fig. 1), including novel or emerging or divergent pathogens. In the case of many viral pathogens evolution and mutations over time can reduce detection with specific PCR reactions. Mutations affecting primer binding sites may reduce binding affinity during the reaction and, for this reason, the performance of diagnostic assays must be monitored closely and at times altered. NGS could, therefore, be used as an adjunct in the quality control of PCR assays.
It is possible to detect full genome sequences from diagnostic samples and even with partial genome sequence it is feasible to subtype viral pathogens. Real-time knowledge of the circulating viral subtype is of particular importance in the management of influenza where this informs anti-viral choice, potential resistance and vaccine efficacy. This is currently carried out using additional PCR assays and Sanger sequencing, although this is not always possible in real-time.
Laboratory workflow
Currently the identification of rare or unusual pathogens using molecular methods necessitates samples to be batched to make the process cost effective; alternatively the test is centralized to a single laboratory to which samples must be sent. Either results in an increase in turn-around time. The use of NGS without any enrichment or targeting would permit samples to be treated in the same manner irrespective of type or likely pathogen.
Challenges
A major barrier in introducing NGS to the diagnostic setting is cost. Although the cost of NGS is decreasing rapidly it remains considerably more expensive than multiplex PCR. It is, therefore, unlikely to be cost effective to use this method for pathogen detection in non-severe infections for the time being. However, any cost–benefit analysis on introducing NGS to a diagnostic setting should also consider, on the positive side of the balance sheet, the likely savings NGS would offer in reductions to epidemiological and public health testing.
Complexity and turnaround time
Current methods of library preparation are complex requiring multiple user interventions and additional equipment to that found in a diagnostic laboratory with attendant implications for the time and cost of the process. To be carried out as a routine diagnostic assay these processes would need to be simplified and, ideally, automated to reduce hands-on time and the potential for contamination and human error.
The commonly used sequencing platforms take several hours or even days to generate sequence information. It should be noted that the third generation platforms that use single-molecule real-time (SMRT) technology are rapid and, as the name suggests, can be analysed in almost real-time.
Data analysis
Data analysis and storage is a major bottleneck in the NGS process. The computational power required for analyses would be beyond the current capabilities of diagnostic services. The methods used in data analysis pose a further challenge. Currently there is no agreed method as to the best approach for data analysis; indeed this is an entire specialty in itself, bioinformatics. Development of software programmes will both make the analysis more feasible in a diagnostic service to non-bioinformaticians and will lead to standardization of data processing.
Discussion
NGS undoubtedly has potential to dramatically change the landscape of infection diagnostics. Whether it will replace current molecular methods remains to be seen. The cost and complex sample processing remains prohibitive but these novel technologies are still in an exponential phase of development. Even current methodologies are yielding promising results in this field. The lack of pathogen targeting means that there is potential for a single work flow to be applied to all specimens, no matter what the syndrome which could even be extended to non-viral pathogens, resulting in a pan-microbial diagnostic test.
The generation of virus sequence as part of a diagnostic assay has substantial management and epidemiological benefits. In terms of respiratory infections this is currently limited to resistance testing and strain analysis of influenza. However, in the management of blood-borne viruses, particularly HIV and hepatitis C virus (HCV), point mutations and minor populations may impact greatly on the management and prognosis of patients. With the introduction of novel therapies or vaccines against viral respiratory infections NGS will have an even greater clinical benefit.
Acknowledgements
I would like to thank Dr Rory Gunson and Dr Emma Thomson for reviewing the manuscript.
References
1. Clark TW, Medina MJ, Batham S, Curran MD, Parmar S, Nicholson KG. Adults hospitalised with acute respiratory illness rarely have detectable bacteria in the absence of COPD or pneumonia; viral infection predominates in a large prospective UK sample. J Infect 2014; 69(5): 507–515.
2. Linares J, Ardanuy C, Pallares R, Fenoll A. Changes in antimicrobial resistance, serotypes and genotypes in Streptococcus pneumoniae over a 30-year period. Clin Microbiol Infect 2006; 16(5): 402–410.
3. Lindbaek M. Prescribing antibiotics to patients with acute cough and otitis media. Br J Gen Pract 2010; 56(524): 164–166.
4. Butler CC, Hood K, Verheij T, Little P, Melbye H, Nuttall J, Kelly MJ, Mölstad S, Godycki-Cwirko M, Almirall J, Torres A, Gillespie D, Rautakorpi U, Coenen S, Goossens H. Variation in antibiotic prescribing and its impact on recovery in patients with acute cough in primary care: prospective study in 13 countries. BMJ 2009; 338: b2242.
5. van Elden LJ, van Kraaij MG, Nijhuis M, Hendriksen KA, Dekker AW, Rozenberg-Arska M, van Loon AM. Polymerase chain reaction is more sensitive than viral culture and antigen testing for the detection of respiratory viruses in adults with hematological cancer and pneumonia. Clin Infect Dis 2002; 34(2): 177–183.
6. FTD Respiratory Pathogens 33. Fast-track Diagnostics 2016. (http://www.fast-trackdiagnostics.com/products/ftd-respiratory-pathogens-33/)
7. Nickbakhsh S, Thorburn F, von Wissmann B, McMenamin J, Gunson RN, Murcia PR. Extensive multiplex PCR diagnostics reveal new insights into the epidemiology of viral respiratory infections. Epidemiol Infect 2016; 144(10): 2064–2076.
8. Thorburn F, Bennett S, Modha S, Murdoch D, Gunson R, Murcia PR. The use of next generation sequencing in the diagnosis and typing of respiratory infections. J Clin Virol 2015; 69: 96–100.
9. Prachayangprecha S, Schapendonk CM, Koopmans MP, Osterhaus AD, Schürch AC, Pas SD, van der Eijk AA, Poovorawan Y, Haagmans BL, Smits SL. Exploring the potential of next-generation sequencing in detection of respiratory viruses. J Clin Microbiol 2014; 52(10): 3722–3730.
The author
Fiona Thorburn PhD
NHS Greater Glasgow and Clyde, Glasgow G12 0XH, UK
E-mail: Fionathorburn@nhs.net