Next-generation sequencing in clinical diagnostics and genomics research
The UK prime minister recently announced an investment package worth £300 million pounds for genomic research. This will include the sequencing of 100,000 genomes by 2017. The project, driven by Genomics England, will have a major impact on many areas of healthcare. Next-generation sequencing (NGS) technology is the method by which this sequencing will be achieved. NGS is currently being used in many healthcare services.
by Dr K. Gilmour
Background
Sequencing of the first human genome took 10 years to complete at a cost of USD300 billion. Although genomics has been recognized and hailed as the future of medicine, the costs associated with sequencing were considered prohibitive. Scientists proposed that large-scale projects would be required to decipher the secrets within each genome and how they interconnect with disease susceptibility, progression and treatment. In 2005 next-generation sequencing (NGS) became commercially available and in the 9 years since has transformed genomics beyond all recognition. Large-scale projects are now financially feasible and the potential of genomics and its link with healthcare can finally be realized.
Different NGS technologies are commercially available with Illumina and Ion torrent™ (Life Technology) probably considered the market leaders. Some NGS instruments can generate a terabase of sequence data in a single run. This equates to around 500 human genomes a week, each costing near to the USD1000 mark in reagents, a financial figure hailed as the ultimate goal. NGS is faster, more accurate and much more sensitive than traditional Sanger sequencing and will contribute directly to improvements in diagnostic medicine, personalized medicine and medical research.
An overview of NGS technology
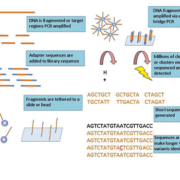
The details of the NGS workflow differ from technology to technology but the main principle remains the same. Extracted DNA from human, animal or microbe sources, is turned into a ‘library’ of DNA. This usually involves making the large pieces of DNA smaller (fragmenting) and then adding special handles known as ‘adapter DNA’ to the ends of each of the DNA fragments (Fig. 1). Adapters are merely small pieces of DNA of known sequence, which can be used to manipulate the fragments of DNA in order to sequence them. This manipulation includes tethering the individual fragments to either a slide or a tiny bead onto which the fragment is clonally amplified producing millions of DNA molecules all of the same sequence. The whole library of different clonally amplified fragments is then sequenced simultaneously. NGS sequencing chemistry produces a detectable ‘signal’. This signal is often fluorescent, so each time a single nucleotide (A, G, C or T) is incorporated into a DNA molecule a tiny amount of light is emitted and detected. The individual sequence produced is known as a ‘read’ and once the millions of small reads in the reaction have been generated they are aligned and assembled via computer algorithms into much longer sequences. Because millions of reads are generated even molecules of low abundance can be sampled making this technique extremely sensitive. Large sequencers able to generate hundreds of human genome sequences a week can be used in high-throughput research projects. Small, fast bench-top sequencers are also available and are highly suited to the demands of a clinical laboratory.
Human genomics
Identifying the genes involved in rare disorders can help doctors to diagnose and understand the underlying cause and nature of the disease and in turn determine what treatment a patient requires. Genomics offers a global look at all genes and how they interact instead of focusing on specific genes and biochemical pathways. Sequencing the exomes (the parts of the genome that encode genes) of only a few people with a rare genetic disorder can locate the mutated gene involved [1]. Genome-wide association studies (GWAS) are also allowing researchers to identify genes associated with many common diseases and so they help predict how likely people are to suffer from specific diseases in their life-time including such things as Parkinson’s disease [2].
NGS in non-invasive prenatal diagnosis
The sensitivity of NGS makes it ideal for non-invasive prenatal diagnosis of fetal aneuploidies. Maternal blood often contains cell-free fetal DNA at very low concentrations. NGS can be used to pick up anomalies in this DNA and so a simple blood test can replace invasive techniques [3].
Personalized medicine
The ability to stratify patient responses to drugs based on the individual’s genetic content has revolutionised how drug trials are performed and the speed at which new drugs reach the manufacturing stage. In cancer medicine, determining the genetic profile of a patient’s tumour can predict which drugs the tumour will potentially respond to thus reducing the likelihood of exposure to a drug with terrible side effects and no clinical benefit [4]. Currently, tumours of many cancer types are regularly tested for individual gene mutations, the results of which determine the treatment. As research reveals further biomarkers of drug response, multiple genes will need to be tested. It is no longer cost effective to test for each of these biomarkers individually and NGS offers the ability to sequence all or part of the tumour genome. The sensitivity of NGS allows mutations to be detected in tissue that contains only a small number of tumour cells. In most hospitals tumour tissue is formalin fixed and embedded in paraffin (FFPE) before being section and mounted on slides for histopathology review. This process can often lead to DNA damage, including fragmentation, rendering the DNA useless for some molecular techniques. As NGS relies on short DNA fragments, FFPE extracted DNA can still be used [5].
NGS in microbiology
In order to prescribe the correct anti-retroviral drugs, the resistance genes of the HIV strain a patient carries are often sequenced. Sanger sequencing would require 20% of the HIV viral population to contain the drug resistance gene in order to be detected. ‘Deep sequencing’ or sequencing the genome many times using NGS can detect resistance genes even if present in less than 1% of the viral population [6]. Outbreaks of dangerous Escherichia coli strains can now be detected early and spread prevented because of the speed at which the sequencing and reconstruction of the relationships of the isolated strains can be achieved [7]. NGS continues to grow as the technology of choice in microbiology.
Possible problems with NGS
With any new technology or venture on the scale of the Genomics England ‘100,000 Genomes Project’ there are potential problems.
Data analysis
The availability of small bench top sequencers means that even small diagnostic labs will be able to use NGS. Different NGS platforms generate different types of data with differing degrees of quality. Because of the inherent errors of enzymatic driven sequencing and the variability in the sequencing signals generated, a host of clever computer algorithms are needed to determine the likelihood of every base in the sequence being correct. The algorithms used to do these analyses are often sold packaged as software or analysis pipelines and are designed by in-house bioinformaticians. With the misinterpretation of sequence data carrying such dire consequences, robust data analysis is paramount. Illumina will be the technology used for all the sequence data generated by the 100,000 Genome Project so all data will likely be handled, processed and analysed in a very similar manner leading to reproducible and robust results. Other clinical laboratories entering into the sequencing revolution will be bombarded with options of technology as well as analysis methods. Clinical laboratories in most countries adhere to a set of rigorous assessments and standards and all clinical tests must be fully validated. Validation of NGS is complicated but best practice guidelines are aiming to simplify the process. ‘Targeted sequencing’, where panels of only a few to a few hundred clinically relevant genes are sequenced makes validation and analysis easier. Unifying analysis processes will remain an important consideration in the future.
Data storage and security
The 100, 000 Genome Project will produce petabytes of data, but even small diagnostic labs will be producing large quantities of data. Targeted gene panels will help but data storage could still be an issue. NGS generates sequence files and associated raw data files and deciding what should be stored and discarded is debated. The Royal College of Pathologists guidelines recommend that data and records pertaining to pathology tests be retained for a minimum of 25 years. DNA sequence is of a highly sensitive nature as even without patient details attached, it contains all the information to link it the individual from which it was taken. Secure storage of DNA sequence with compression and encryption is an important consideration. The Medical Research Council in the UK has earmarked £24 million pounds of the Genomics England funding for computing power, including analysis and secure storage.
Ethical implications
The mainstream adoption of any new technology has ethical implications. Whilst sequencing a patient’s tumour to determine a cancer treatment plan another gene mutation could be identified, unrelated to the condition being treated. In the UK all patients must consent to any germ-line genetic test. Genetic counselling is offered and patients are helped to come to terms with the implications of the findings. Serendipitous discoveries have the potential to create many ethical dilemmas for clinicians.
The future: a learning healthcare system
Although powerful, medical genomics has so far not had the major impact on healthcare predicted at the time of the release of the first human genome. The 100,000 Genome Project will change that. The project hopes to link up genomic data with the medical records for each patient. This means that research data can be actively generated as the project persists. Every person consenting to the project will be a walking research project from which we can learn important lessons about treatment and response [8]. This could transform our UK healthcare system into a learning environment like no other in the world. It will generate the evidence on which future improvements can be made. With strong collaborative partnerships set up with Illumina, the Wellcome Trust Sanger institute, Medical Research Council, and Cancer Research UK to name but a few, this the Genomics England project has the potential to be a great success.
So-called ‘third generation sequencing’ technology is already a reality and NGS sequencing chemistries are continually evolving and improving. Although it is unlikely in the very near future that every person in the country will have their genome sequenced, NGS is still contributing massively to healthcare improvements in genomics and other clinical diagnostic areas.
References
1. Boycott KM, Vanstone MR, Bulman DE, MacKenzie AE. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat Rev Genet. 2013; 14(10): 681–691.
2. Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, DeStefano AL, Kara E, Bras J, et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat Genet. 2014; doi: 10.1038/ng.3043. [Epub ahead of print].
3. Nepomnyashchaya YN, Artemov AV, Roumiantsev SA, Roumyantsev AG, Zhavoronkov A. Non-invasive prenatal diagnostics of aneuploidy using next-generation DNA sequencing technologies, and clinical considerations. Clin Chem Lab Med. 2013; 51(6): 1141–1154.
4. Jackson SE, Chester JD. Personalised cancer medicine. Int J Cancer 2014; doi: 10.1002/ijc.28940. [Epub ahead of print].
5. Fairley JA, Gilmour K, Walsh K. Making the most of pathological specimens: molecular diagnosis in formalin-fixed, paraffin embedded tissue. Curr Drug Targets 2012; 13(12): 1475–1487.
6. Gibson RM, Schmotzer CL, Quiñones-Mateu ME. Next-generation sequencing to help monitor patients infected with HIV: ready for clinical use? Curr Infect Dis Rep. 2014; 16(4): 401.
7. Veenemans J, Overdevest IT, Snelders E, Willemsen I, Hendriks Y, Adesokan A,Doran G, Bruso S, Rolfe A, Pettersson A, Kluytmans JA. Next-generation sequencing for typing and detection of resistance genes: performance of a new commercial method during an outbreak of extended-spectrum-beta-lactamase-producing Escherichia coli. J Clin Microbiol. 2014; 52(7): 2454–2460.
8. Ginsburg G. Medical genomics: gather and use genetic data in health care. Nature 2014; 508(7497): 451–453.
The author
Katelyn Gilmour PhD
Molecular Pathology, Dept. Laboratory Medicine, Royal Infirmary of Edinburgh, Edinburgh EH16 4SA, UK
*Corresponding author
E-mail: Katelyn.gilmour@nhslothian.scot.nhs.uk