Pharmacogenomics: implications for drug safety
The increased burden of hospital admissions due to adverse drug reactions (ADRs) carries significant implications for patients and healthcare systems. Understanding the correlations between genetics and drug safety may improve clinical outcomes through the realization of personalized medicine. This article outlines a practical approach to pharmacogenomics with examples in clinical practice.
by Dr Marcin Bula and Prof. Sir Munir Pirmohamed
Introduction
The World Health Organization (WHO) defines an adverse drug reaction (ADR) as a response to a drug which is harmful and unintended and which occurs at doses normally used in man for prophylaxis, diagnosis or treatment of disease or the modification of physiological function.
There are several classifications used to describe ADRs taking into account severity, source of reported data, reaction time and location of reaction. In this article, we focus on the most-widely used classification and divide ADRs into two major groups: dose-related (type A – ‘Augmented’) and apparently non-dose-related (type B – ‘Bizarre’). Type A reactions are predictable, more common and usually less serious. They can be managed by simply reducing the dose or withholding the drug. Type B reactions are uncommon, unpredictable and usually more serious. They may either be immunologic or non-immunologic in nature, and because we do not understand pathogenesis, this makes the reactions more difficult to predict and prevent.
The overall incidence of ADR-related hospital admissions is approximately 6.5% [1, 2] although this figure might be an underestimate due to complexity of cases presenting to hospitals, compounded in real-world settings, by the poor reporting of ADRs by healthcare professionals. A previous systematic review of drug-related hospital admissions showed that antiplatelets, NSAIDs and anticoagulants were responsible for more than 50% of the total ADR-related hospitalizations [3]. It has been estimated that ADRs cost the UK National Health Service (NHS) approximately £1 billion annually, and studies in the USA have suggested that ADRs are the fourth to sixth leading cause of death [4].
Type B adverse drug reactions
Type B ADRs are a major concern for healthcare because of their unpredictable multifactorial nature, and potentially life threatening clinical outcomes. The most common organs affected are the skin, liver and blood cells. Some type B ADRs have been found to have a genomic component; the most striking example is the association between abacavir hypersensitivity and human leukocyte antigen (HLA). Abacavir is a guanosine analogue used in combination therapy with other antiretroviral medications in the treatment of human immunodeficiency virus (HIV). Previous studies have shown that approximately 4–8% [5] of patients develop a hypersensitivity reaction (HSR) within the first 6 weeks of treatment, characterized by fever, rash, gastrointestinal symptoms, general malaise, and other less common manifestations, such as headaches, respiratory and musculoskeletal symptoms [6]. The association between abacavir hypersensitivity and the HLA Class I allele, HLA-B*57:01 was first reported in 2002 by two independent research teams in Australia and North America, followed by a study in the United Kingdom. This has been complemented by functional studies that have shown that abacavir hypersensitive HLA-B*57:01 carriers show increased proliferation of CD8+ T lymphocytes following drug exposure. The exact mechanisms underlying the reaction are still not fully understand but in vitro models have shown how abacavir interacts with HLA-B*57:01, and with T cell receptors forming an immunological synapse that results in an immune response. Interestingly approximately 50% patients who are carriers of HLA-B*57:01 do not develop abacavir hypersensitivity, but the reasons for this are unknown. A study in the NHS (UK) showed that genetic testing before abacavir initiation is cost-effective [7]. Both the Food and Drug Administration (FDA) and European Medicines Agency (EMA) recommend screening for HLA-B*57:01 even though the carriage rate varies according to ethnicity from 5–8% in Europeans to 2.4% in African Americans [8]. Pre-prescription genotyping has been shown to be highly cost-effective and has reduced the incidence of abacavir hypersensitivity from over 5% to less than 1%.
It is estimated that epilepsy affects 1% of the population worldwide. Carbamazepine is an aromatic anticonvulsant that is also used for trigeminal neuralgia and bipolar disease. Cutaneous adverse reactions to carbamazepine are wide-ranging, and can manifest as maculopapular eruptions at the mild end, to the more severe cutaneous adverse reactions (which include drug reactions with eosinophilia and systemic symptoms (DRESS), Stevens–Johnson syndrome (SJS) and toxic epidermal necrolysis (TEN). SJS/TEN are the most serious ADRs with mortality rates of 5% for SJS and 35% for TEN [9]. SJS and TEN represent a continuum of cutaneous reactions, with the degree of skin detachment able to differentiate between the two (SJS involves less than 10% of the body surface area, whereas TEN affects more than 30% of the body surface area). A study in 2004 found a strong association between HLA-B*15:02 and SJS induced by carbamazepine in Han Chinese. This has been replicated by many other studies undertaken in Han Chinese, Thai and Malays, and a prospective study by Chen et al. [10] subsequently showed that genetic testing prior to treatment significantly reduced the incidence of carbamazepine-induced SJS. The association with HLA-B*15:02 is limited to South East Asian populations, and has not been demonstrated in Northern Europeans because the population prevalence of this allele is very low (<0.5%). Currently regulatory bodies including the FDA and EMA recommend genotyping for HLA-B*15:02 in South East Asian populations before starting treatment with carbamazepine, although various commentaries have questioned what is exactly meant by a South East Asian population. This reflects the difficulties in assigning screening based on self-reported ethnicity as it does not take into account admixture that occurs in almost all populations, and can exclude populations that may also be susceptible but would not be considered to be South East Asian. There is some evidence to show that HLA-B*15:02 may also predispose to SJS/TEN with phenytoin although the risk estimates are much less than with carbamazepine.
More recently, the HLA-A*31:01 allele, which is common in most ethnic groups has been associated with a range of carbamazepine hypersensitivity phenotypes including DRESS and SJS/TEN. In a Han Chinese population, an association with HLA-A*31:01 and carbamazepine-induced DRESS was demonstrated but not with SJS/TEN (where HLA-B*15:02 is predominant). In terms of mechanisms, it is not clear why HLA-B*15:02 only predisposes to SJS/TEN with carbamazepine, whereas HLA-A*31:01 predisposes to a wider range of phenotypes; cooperativity between different HLA alleles, for example with the HLA-DRB1*04:04, and with T-cell receptor clonotypes may be important in determining the phenotype (11). Genetic testing of HLA-A*31:01 is not mandatory at the moment; however, a UK study has recently shown that genotyping before initiating carbamazepine in the NHS would be cost effective (12).
Type A adverse drug reactions
Two interesting examples of the modern use of pharmacogenomics to prevent type A ADRs are with eliglustat and warfarin. Gaucher’s disease (GD) is the most common lysosomal storage disorder, which is inherited in an autosomal recessive fashion with an incidence of 1 in 40 000–60 000 in the general population, and 1 in 450 in Ashkenazi Jews [13]. Type 1 GD is the most common variant affecting more than 90% of all patients without neurological involvement, opposite to the manifestations observed with types 2 and 3 GD. Reduced activity of the β-glucocerebrosidase enzyme as a result of the GBA gene mutation leads to lysosomal accumulation of undegraded glucosylceramide causing dysfunction of various organs. For the last 20 years, the standard treatment for GD has been enzyme replacement therapy (ERT) requiring twice weekly intravenous infusions with a recombinant form of human β-glucosidase. Eliglustat represents an example of a new therapeutic strategy in GD – substrate reduction therapy (SRT), which is characterized by inactivation of glucosylceramide synthase involved in glucosylation of ceramide [14]. Eliglustat undergoes extensive metabolism by cytochrome P450 enzymes, in particular by CYP2D6 and to a lesser extent by CYP3A4. Studies have confirmed a strong correlation between the CYP2D6 metabolizer status and drug exposure. Eliglustat has recently been approved by both the FDA and EMA for the treatment of patients with type I GD – interestingly, given the strong effect of the CYP2D6 gene polymorphism on drug exposure patients need to be genotyped for their CYP2D6 metabolizer status, and the dose needs to be reduced by 50% in poor metabolizers. Furthermore, co-administration of drugs inhibiting CYP2D6 needs to be prescribed with extreme caution to prevent dose-dependent ADRs.
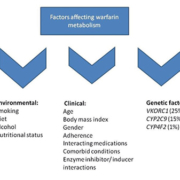
Warfarin is a vitamin K antagonist that is a mainstay of anticoagulation treatment in venous thromboembolism (VTE) and stroke prevention in atrial fibrillation (AF). Vitamin K antagonist therapy (despite high clinical effectiveness) has significant disadvantages and limitations including a narrow therapeutic index, drug and food interactions, routine coagulation monitoring and dose adjustments. Polymorphisms in CYP2C9 and VKORC1 genotypes and inter-individual variability can significantly influence warfarin metabolism and pharmacodynamic (PD), hence the increased risk of significant adverse reaction such as hemorrhage (Fig. 1) [15, 16]. The genetic determinants of warfarin metabolism have been heavily investigated since 1990. CYP2C9 and VKORC1 are the two main genes associated with warfarin dose requirements. Additional genetic variants, such as CYP4F2, contribute to warfarin metabolism; however, their role is less significant. The International Warfarin Pharmacogenetics Consortium proved that, based on previous studies, algorithms incorporating genetics factors (CYP2C9 and VKORC1) are more precise in prediction warfarin dosing algorithms. However, two recent large randomized controlled trials, EU-PACT and COAG, showed conflicting evidence of the role of pharmacogenetics compared to clinically guided warfarin dosing [17]. It is estimated that different outcomes in the EU-PACT and COAG trials are due to various factors including ethnic heterogeneity, genotype information on day one dosing and different control arms. The clinical utility of genotype-based warfarin dosing would need further research in particular in populations other than Caucasians.
Conclusions
Pharmacogenomics is an important area of study in understanding and preventing ADRs. It can be used throughout the whole cycle of drug development. During the pre-clinical stages, determination of how a drug is metabolized and eliminated from the body can provide valuable information on how polymorphisms in drug metabolizing enzymes and transporters affect drug pharmacokinetics and will lead to valuable prescribing information in the summary of product characteristics. This could be followed by specific, subsequent studies that may lead to genotype-dependent dosing, as in the case of eliglustat. Such precise dosing is not commonplace now, but is likely to become more important in the future. Dosing is a key determinant of the risk of ADR, and one that is still ignored. Rare and often more serious ADRs such as hypersensitivity are often not detected until phase IV, and this will require post-marketing studies. This is beautifully exemplified by abacavir hypersensitivity and the different studies that showed an association with HLA-B*57:01.
Implementation of pharmacogenomics into clinical practice has been patchy overall. This is because of many reasons, including poorly replicated gene-drug associations. However, even when the associations have been replicated and are biologically convincing, implementation has sometimes not occurred. This may be because pharmacogenetics (and the whole area of personalized medicine) represents a disruptive innovation that changes the whole clinical pathway. Changing behaviour through re-engineering the clinical pathways in a healthcare setting will require changes in the systems currently employed to deliver clinical care, which can be likened to turning around a supertanker – i.e. it will take time, money and cooperation of every part of the whole healthcare system. Of course, further research is also need in many other areas, and it is important that research in pharmacogenomics is combined with other modalities to ensure that we are covering all possible factors that can affect a response to a drug.
References
1. Pirmohamed M, James S, Meakin S, Green C, Scott AK, Walley TJ, Farrar K, Park BK, Breckenridge AM. Adverse drug reactions as cause of admission to hospital: prospective analysis of 18 820 patients. BMJ 2004; 329(7456): 15–19.
2. Howard RL, Avery AJ, Howard PD, Partridge M. Investigation into the reasons for preventable drug related admissions to a medical admissions unit: observational study. Qual Saf Health Care 2003; 12(4): 280–285.
3. Howard RL, Avery AJ, Slavenburg S, Royal S, Pipe G, Lucassen P, Pirmohamed M. Which drugs cause preventable admissions to hospital? A systematic review. Br J Clin Pharmacol. 2007; 63(2): 136–147.
4. Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA 1998; 279(15): 1200–1205.
5. Peyriere H, Guillemin V, Lotthe A, Baillat V, Fabre J, Favier C, Atoui N, Hansel S, Hillaire-Buys D, Reynes J. Reasons for early abacavir discontinuation in HIV-infected patients. Ann Pharmacother. 2003; 37(10): 1392–1397.
6. Clay PG. The abacavir hypersensitivity reaction: a review. Clin Ther. 2002; 24(10): 1502–1514.
7. Hughes DA, Vilar FJ, Ward CC, Alfirevic A, Park BK, Pirmohamed M. Cost-effectiveness analysis of HLA B*5701 genotyping in preventing abacavir hypersensitivity. Pharmacogenetics 2004; 14(6): 335–342.
8. Cao K, Hollenbach J, Shi X, Shi W, Chopek M, Fernandez-Vina MA. Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum Immunol. 2001; 62(9): 1009–1030.
9. Roujeau JC, Stern RS. Severe adverse cutaneous reactions to drugs. N Engl J Med. 1994; 331(19): 1272–1285.
10. Chen P, Lin JJ, Lu CS, Ong CT, Hsieh PF, Yang CC, Tai CT, Wu SL, Lu CH, Hsu YC, et al. Carbamazepine-induced toxic effects and HLA-B*1502 screening in Taiwan. N Engl J Med. 2011; 364(12): 1126–1133.
11. Lichtenfels M, Farrell J, Ogese MO, Bell CC, Eckle S, McCluskey J, Park BK, Alfirevic A, Naisbitt DJ, Pirmohamed M. HLA restriction of carbamazepine-specific T-Cell clones from an HLA-A*31:01-positive hypersensitive patient. Chem Res Toxicol. 2014; 27(2): 175–177.
12. Plumpton CO, Yip VL, Alfirevic A, Marson AG, Pirmohamed M, Hughes DA. Cost-effectiveness of screening for HLA-A*31:01 prior to initiation of carbamazepine in epilepsy. Epilepsia 2015; 56(4): 556–563.
13. Zeller JL, Burke AE, Glass RM. JAMA patient page. Gaucher disease. JAMA 2007; 298(11): 1358.
14. McEachern KA, Fung J, Komarnitsky S, Siegel CS, Chuang WL, Hutto E, Shayman JA, Grabowski GA, Aerts JM, Cheng SH, Copeland DP, Marshall J. A specific and potent inhibitor of glucosylceramide synthase for substrate inhibition therapy of Gaucher disease. Mol Genet Metab. 2007; 91(3): 259–267.
15. Johnson JA, Cavallari LH. Warfarin pharmacogenetics. Trends Cardiovasc Med. 2015; 25(1): 33–41.
16. Wysowski DK, Nourjah P, Swartz L. Bleeding complications with warfarin use: a prevalent adverse effect resulting in regulatory action. Arch Intern Med. 2007; 167(13): 1414–1419.
17. Pirmohamed M, Kamali F, Daly AK, Wadelius M. Oral anticoagulation: a critique of recent advances and controversies. Trends Pharmacol Sci. 2015; 36(3): 153–163.
The authors
Marcin Bula* MBBS, MRCP(L); Munir Pirmohamed MB ChB (Hons), PhD, FRCP, FRCP(E), FBPhS, FMedSci
Institute of Translational Medicine, University of
Liverpool, Liverpool L69 3GL2, UK
*Corresponding author
E-mail: m.bula@liverpool.ac.uk