Pharmacometabonomics: a new methodology for personalized medicine
The application of metabolic profiling of human biofluids to the prediction of drug efficacy, pharmacokinetics, metabolism and/or toxicity forms a paradigm known as pharmacometabonomics. Pharmacometabonomics holds out promise for the improved delivery of personalized medicine in the future, as it takes into account both genetic and environmental factors, including diet, drug intake and most notably, the status of the gut microbiome, in deriving predictions. Pharmacometabonomics is thus complementary to pharmacogenomics and in some instances the two technologies can be synergistically used together. This article introduces pharmacometabonomics and covers current important application areas.
by Dr Dorsa Varshavi, Dorna Varshavi and Prof. Jeremy Everett
Introduction
In 21st century medicine, a major goal is to develop personalized medicine for selected groups of patients in order to reduce the likelihood of adverse drug reactions and to maximize the desired therapeutic effect [1]. Until recently, personalized drug therapy was delivered almost exclusively by pharmacogenomics (PG), where an individual’s genetic makeup is used to predict the outcome of drug treatment [2]. The best-recognized examples of PG involve drug effect predictions made by analysis of genetic polymorphisms in drug-metabolizing enzymes such as the cytochrome P450 isoenzymes [3].
Although genetic variation is an important determinant of individual variability in drug response, it is now well recognized that personalized drug therapy cannot always be attained using genetic knowledge alone. This is because inter-individual variation in drug response is a consequence of multiple factors, including genetic and epigenetic factors and in addition, environmental factors such as nutritional and health status, the condition of the microbiome, exposure to environmental toxins, and co- or pre-administration of other drugs, including alcohol. These environmental factors can strongly affect drug absorption, distribution, metabolism and excretion and thereby cause inter-individual variation in drug efficacy and safety.
Metabonomics is defined as: ‘The study of the metabolic response of organisms to disease, environmental change, or genetic modification’ [4]. In a metabonomics experiment, changes in the levels of biofluid or tissue metabolites, before and after an intervention, such as drug administration, are measured using analytical technologies such as nuclear magnetic resonance (NMR) spectroscopy or mass spectrometry (MS). The alternative term metabolomics is also used and although its definition is observational, rather than interventional, the two terms are now used interchangeably.
Pharmacometabonomics is a recent development from metabonomics and is defined as ‘the prediction of the outcome (for example, efficacy or toxicity) of a drug or xenobiotic intervention in an individual based on a mathematical model of pre-intervention metabolite signatures’ [5–7]. In contrast to a metabonomics experiment, where the effect of an intervention is assessed based on changes in metabolite profiles post-intervention, in a pharmacometabonomics experiment the effect of the intervention is predicted based on the pre-intervention metabolite profiles.
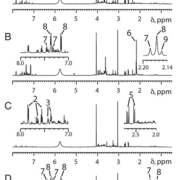
Although first demonstrated in animals [5], pharmacometabonomics was quickly also demonstrated in humans, in a study where an individual’s pre-dose urinary endogenous metabolite profile was used to predict the metabolism of the analgesic paracetamol [6]. NMR-based analyses showed that individuals excreting relatively high levels of the microbial co-metabolite para-cresol sulfate in their pre-dose urine, excreted less paracetamol sulfate and more paracetamol glucuronide post-dose than individuals excreting low pre-dose amounts of para-cresol sulfate (Figs 1 & 2). Para-cresol sulfate is a metabolite produced from the hepatic sulfation of para-cresol, which is itself generated by gut bacteria, particularly Clostridium species. Paracetamol and para-cresol have similar molecular structures and both compete for limited human sulfation capacity via the same sulfotransferase enzymes, particularly SULT1A1. Thus individuals with a microbiome producing large amounts of para-cresol use up a large degree of their sulfonation capacity in metabolizing this toxin to para-cresol sulfate, and a subsequent dose of paracetamol will be metabolized to a greater degree by glucuronidation. This study was important for two key reasons: (1) it was the first demonstration of pharmacometabonomics in humans and (2) it was the first demonstration of the influence of the gut microbiome on human drug metabolism; the fact that the key biomarker in this study of human drug metabolism was a bacterially-derived molecule was a shock. Furthermore, the findings of this study will have implications for other drugs for which sulfonation is important, as well as certain diseases such as autism where abnormal paracetamol metabolism has been observed.
Pharmacometabonomics has been also used to predict individual responses to therapy, including the prediction of patient responses to treatment with the statin simvastatin. Statins can reduce low-density lipoprotein cholesterol (LDL-C) and, therefore, are used for treatment of cardiovascular disease. Kaddurah-Daouk et al. demonstrated that pre-dose plasma levels of the phosphatidylcholine metabolite PC18:2n6, the cholesterol ester CE18:1n7 and the free fatty acid FA18:3n3, were positively correlated with the magnitude of simvastatin-induced reduction in LDL-C, in 36 good responders and 36 poor responders [8]. A targeted pre-dose plasma analysis by MS then demonstrated (amongst other results) a strong correlation between the degree of reduction in LDL-C and higher pre-dose concentrations of three secondary, bacteria-derived, bile acids: lithocholic acid (LCA), taurolithocholic acid (TLCA) and glycolithocholic acid (GLCA), as well as coprostanol (COPR) [9]. This study further supported the contribution of the microbiome in influencing drug responses.
Pharmacometabonomics studies can be pursued jointly with, or followed up by pharmacogenetics studies. A good exemplification of the so-called ‘pharmacometabonomics informed pharmacogenomics approach’ is an MS-based study which demonstrated that pre-dose plasma levels of glycine, a central nervous system inhibitory neurotransmitter, were associated with rates of response or remission during citalopram/escitalopram treatment in patients with major depressive disorder (MDD) in the Mayo Clinic–NIH Pharmacogenetics Research Network (PGRN) Citalopram/Escitalopram Pharmacogenomics (Mayo–PGRN SSRI) study [10]. Tag single-nucleotide polymorphism (SNP) genotyping of the genes encoding enzymes in the glycine pathway from 529 patients enrolled in the Mayo-PGRN SSRI study was then completed. A series of SNPs in the gene encoding glycine dehydrogenase (GLDC) were found to be significantly associated with disease remission, with rs10975641 SNP showing the strongest association. This study demonstrated that pharmacometabonomics data can inform and complement pharmacogenomics data and, when combined, they can provide improved insights into the mechanisms influencing variability in drug response.
There are now many examples of the use of pharmacometabonomics for the prediction of human drug efficacy, toxicity, metabolism and pharmacokinetics and recent reviews are available [7, 11].
Conclusion and future prospect
Since its initial discovery [5], pharmacometabonomics has been increasingly applied in both preclinical and clinical studies to predict drug safety, efficacy, metabolism and pharmacokinetics. Pharmacometabonomics has an important advantage over pharmacogenomics in that it takes into account both genetic and environmental influences on drug administration. Pharmacometabonomics is itself just one specific example of a broader class of approaches known as predictive metabonomics, where the analysis of pre-intervention metabolite profiles can be used to predict clinical responses to other types of intervention, including diet, exercise, or even just the passage of time. A good example of predictive metabonomics can be seen in the recent study by Wang-Sattler et al. [12], who demonstrated that low baseline levels of glycine and lysophosphatidylcholine were predictive of the development of impaired glucose tolerance and/or type-2 diabetes in hundreds of subjects from the population-based, Cooperative Health Research in the Region of Augsburg (KORA) cohort. Another emerging area for which pharmacometabonomics holds promise is in the monitoring of patients over time as they progress through therapies such as cancer chemotherapy or surgery, a paradigm called longitudinal pharmacometabonomics [13]. This approach involves the metabolic profiling of patients before, during and after clinical therapy, in order to predict responses to future treatments and thus choose the optimal treatment regime. PG is now over 50 years old and is still limited in its impact on the practice of medicine. Pharmacometabonomics is much younger and it will take time for it to impact in the clinical arena. We predict that in the near future, personalized medicine will be conducted with assistance from both PG and pharmacometabonomics.
References
1. Pokorska-Bocci A, Stewart A, Sagoo GS, Hall A, Kroese M, Burton H. ‘Personalized medicine’: what’s in a name? Personalized Medicine 2014; 11(2): 197–210.
2. Jorgensen JT. A challenging drug development process in the era of personalized medicine. Drug Discov Today 2011; 16(19–20): 891–897.
3. Pirmohamed M. Personalized pharmacogenomics: predicting efficacy and adverse drug reactions. Ann Rev Genomics Hum Genet. 2014; 15: 349–370.
4. Lindon J, Nicholson J, Holmes E, Everett J. Metabonomics: Metabolic processes studied by NMR spectroscopy of biofluids. Concepts Magn Reson. 2000; 12(5): 289–320.
5. Clayton T, Lindon J, Cloarec O, Antti H, Charuel C, Hanton G, Provost JP, Le Net JL, Baker D, Walley RJ, Everett JR, Nicholson JK. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature 2006; 440(7087): 1073–1077.
6. Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci U S A 2009; 106(34): 14728–14733.
7. Everett JR. Pharmacometabonomics in humans: a new tool for personalized medicine. Pharmacogenomics 2015; 16(7): 737–754.
8. Kaddurah-Daouk R, Baillie RA, Zhu HJ, Zeng ZB, Wiest MM, Nguyen UT, Watkins SM, Krauss RM. Lipidomic analysis of variation in response to simvastatin in the Cholesterol and Pharmacogenetics Study. Metabolomics 2010; 6(2): 191–201.
9. Kaddurah-Daouk R, Baillie RA, Zhu H, Zeng ZB, Wiest MM, Nguyen UT, Wojnoonski K, Watkins SM, Trupp M, Krauss RM. Enteric microbiome metabolites correlate with response to simvastatin treatment. PLoS One 2011; 6(10): e25482.
10. Ji Y, Hebbring S, Zhu H, Jenkins GD, Biernacka J, Snyder K, Drews M, Fiehn O, Zeng Z, Schaid D, Mrazek DA, Kaddurah-Daouk R, Weinshilboum RM. Glycine and a glycine dehydrogenase (GLDC) SNP as citalopram/escitalopram response biomarkers in depression: pharmacometabolomics-informed pharmacogenomics. Clin Pharmacol Ther. 2011; 89(1): 97–104.
11. Everett JR. NMR-based pharmacometabonomics: a new approach to personalized medicine. In: Everett JR, Harris RK, Lindon JC, Wilson ID. (eds) NMR in Pharmaceutical Sciences, pp 359–372. Wiley 2015.
12. Wang-Sattler R, Yu Z, Herder C, Messias AC, Floegel A, He Y, Heim K, Campillos M, Holzapfel C, Thorand B, Grallert H, Xu T, et al. Novel biomarkers for pre-diabetes identified by metabolomics. Mol Syst Biol. 2012; 8: 615.
13. Nicholson JK, Everett JR, Lindon JC. Longitudinal pharmacometabonomics for predicting patient responses to therapy: drug metabolism, toxicity and efficacy. Expert Opin Drug Metab Toxicol. 2012; 8(2): 135–139.
The authors
Dorsa Varshavi PhD, Dorna Varshavi MSc, Jeremy Everett* PhD
Medway Metabonomics Research Group, University of Greenwich, Chatham, Kent ME4 4TB, UK
*Corresponding author
E-mail: j.r.everett@greenwich.ac.uk