Point-of-care platforms, genetic assays and wireless connectivity
Nucleic acids, which are among the best signatures of disease and pathogens, have traditionally been measured in centralised screening facilities using expensive instruments. Such tests are seldom available on point-of-care (POC) testing platforms. Advancements in simple microfluidics, cellphones and low-cost devices, isothermal and other novel amplification techniques, and reagent stabilisation approaches are now making it possible to bring some of the assays to POCs. This article highlights selected advancements in this area.
by Dr Robert Stedtfeld, Maggie Kronlein and Professor Syed Hashsham
Why point-of-care diagnostics?
Point-of-care diagnostics (POCs) bring selected capabilities of centralised screening to thousands of primary health care centres, hospitals, and clinics. Quick turnaround time, enhanced access to specialised testing by the physicians and patients, sample-in-result-out capability, simplicity, ruggedness and lower cost are among the leading reasons for the emergence of POCs. Another advantage of POCs is its flexibility to be adopted for assays that have received less attention and therefore are often “home brewed”, meaning an analyst develops it within the screening facility for routine patient care. The societal benefit–cost analysis of POCs may often exceed the traditional approaches by 10- to 100-fold. However, POCs must deliver the same quality of test results that is available with the existing centralised screening. Centralised screening is well established, has a performance record and analytical expertise ensuring reliability. POCs are emerging and, therefore, for successful integration into the overall healthcare system, POCs must provide an advantage over the existing system consisting of sample transport to a centralised location followed by analysis and reporting. Besides answering why POCs are better than the existing approaches, they must face validation and deployment challenges.
On the positive side, POCs are expected to have lower financial and acceptance barriers compared to what is faced by more expensive traditional approaches because of the need for lowering the cost of diagnostics in general. In 2011, the global in vitro testing market was $47.6 billion and projected to be $126.9 billion by 2022 (http://www.visiongain.com/). At present POCs constitute approximately one third of the total market – distributed in cardiac markers (31%), HbA1c (21%), cholesterol/lipids (16%), fecal occult blood (14%), cancer markers 98%), drug abuse (4%), and pregnancy (4%). Market forces critically determine the pace of technical development and deployment of POCs. Consider, for example, the global market for blood sugar testing (examples for genetic assays on POCs are non-existent) that is estimated to be $18 billion by 2015 and the alternative test, A1c that is only $272 million in 2012. Even though, A1c testing is now indispensable in managing diabetes, it has not received the priority it deserves due to much lower frequency of testing and therefore smaller market. Lowering the cost further, makes its deployment and diffusion even more challenging. Thus POCs must tackle the inherent bottleneck in their business model, i.e. how to succeed with an emerging or new technology, priced to be low cost, but without the access to market and high sales volumes – at least initially.
One option is to use the existing network of cellphones as one component of the POCs. Diagnostic tools based on cellphones and mobile devices have the potential to significantly reduce the economic burden and play an important role in providing universal healthcare. By 2015 the number of cellphone users will reach 1.4 billion and at least 500 million of them will have used health related applications (mHealth) in some form. Currently, more than 17,000 mHealth apps are available on various platforms. However, their ability to carry out genetic assays is yet to be harnessed. Out of the more than 2,500 genetic assays available, perhaps none are available on a mobile platform (GeneTests: www.genetests.org/). The coming decade is predicted to merge genomics, microfluidics and miniaturisation and multiply its impact many-fold by leveraging the resources and cellphone networks. Such platforms may allow the possibility of establishing an open source model for assays that are commercially not viable due to very low volumes.
A key question and the focus of this article is can genetic assays that are currently possible only in centralised screening facilities be carried out on POC platforms? We believe that through a combination of emerging molecular techniques, low-cost simple microfluidic systems, and some additional developments in detection systems and information transfer, it is possible to carry out genetic assays including mutation detection on POCs within the next 5 years and possibly sequencing within a decade.
Existing POC-adaptable genetic technologies
Nucleic acid-based amplification techniques remain the widely used analytical technique for genetic diagnostics. However, integrated systems capable of reliable detection with sensitivity and specificity required for clinical applications are still scarce. In centralised screening facilities, quantitative polymerase chain reaction (qPCR) is the workhorse for genetic analyses. Compared to qPCR, isothermal amplification strategies have been recognised as a promising alternative especially for POCs. This is because of the complexity of establishing the temperature cycling for qPCR and detection systems in POC devices. The advantages of isothermal amplification include high amplification yields (in some instances allowing a positive reaction to be observed with the naked eye), savings in power consumption without the need for temperature cycling, and low time to a positive amplification (as low as 5 minutes for larger copy numbers). Many isothermal techniques have been developed [1] including: loop-mediated isothermal amplification (LAMP), recombinase polymerase amplification (RPA), nucleic acid sequence-based amplification (NASBA), smart amplification process (SmartAmp), rolling circle amplification (RCA), multiple displacement amplification (MDA), helicase-dependent amplification (tHDA), strand displacement amplification (SDA), isothermal and chimeric primer-initiated amplification (ICAN), cross-priming amplification (CPA), single primer isothermal amplification (SPIA), self-sustained sequence replication reaction (3SR), transcription mediated amplification (TMA), genome exponential amplification reaction (GEAR) and exponential amplification reaction (EXPAR).
The benefits of one isothermal technique over another will depend on the application of interest. Techniques requiring a large number of enzymes and that are carried out at low temperature may be less amenable to POCs than those that require a single enzyme. More than one enzyme may, in general, increase the cost, rigor and complexity of the amplification reaction in a POC. While larger number of primer sets will increase the specificity, they will also make the design of primers to target a certain phylogenetic group or divergent functional gene more difficult, if not impossible. This is because of the need for multiple target specific regions, each being a certain distance (number of bases) between the other, and the increased complexity when trying to incorporate degenerate bases in multiple primer sequences within an assay. Isothermal assay enzymes that work at low temperature (less than 40°C) may have a disadvantage in hot and warm climatic conditions. However, an isothermal amplification strategy that directly incorporates primers/probes designed for previously validated qPCR assays, uses a single enzyme, can be performed at higher temperatures, and allows for accurate quantification, will greatly increase the attraction of isothermal amplification, ushering in a new era of point of care genetic diagnostics. The cost associated with licensing an amplification technique will also dictate if it can be used for POCs applications, specifically in low resource settings.
Existing POC platforms for genetic analysis
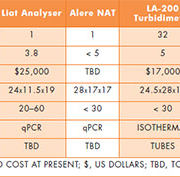
Multiple platforms have been developed for POC genetic testing with an emphasis on reduced costs, sizes, throughput, accuracy and simplicity. Table 1 is a non-exhaustive list to illustrate some of the capabilities. Ideally, POCs must simplify the genetic analysis by accepting crude or unprocessed samples. All of the listed qPCR platforms automatically perform sample processing (cell lysing and DNA purification) directly within the cartridge that the sample is dispensed. Compared to qPCR POCs, isothermal assay POCs have not focused as much on sample processing. There are two reasons for this. One, isothermal assays are generally less influenced by sample inhibitors and may not even require it in certain cases. Second, development of POCs based on isothermal assays has lagged because the assays themselves are relatively new for the diagnostics application.
Development of isothermal genetic POC devices, however, is relatively easier compared to qPCR devices. This is because isothermal genetic POCs utilise components that are inexpensive, smaller and have less power consumption. Use of such components is possible due to the high product yields of isothermal amplification techniques. LAMP, for example, produces 10 µg of DNA in a 25 µl volume compared to 0.2 µg in PCR. This high yield can be quantified with less sophisticated optics compared to those used in qPCR devices. The Gene-Z platform [figure 1], for example, uses an array of individually controlled low power light emitting diodes for excitation and optical fibres (one for each reaction well) for channelling the excitation light to a single photodiode detector for real time measurement [2].
Although POCs are generally considered as a single-assay device, multiplexing of targets (e.g. in co-infections) and analysing a given pathogen with greater depth (e.g. methicillin resistance Staphylococcus aureus, or HIV genotyping) is becoming absolutely critical. Genetic analysis is expected to allow resolution of genotype that is better than that possible by immunoassays. Use of simpler but powerful microfluidic chips (e.g. used with Gene-Z or GenePOC) instead of conventional Eppendorf tubes can be advantageous in terms of cost and power of analysis. Such microfluidic chips are increasingly changing their shape, form, and material and are bound to be simpler, better and more accessible. An example is the paper-based diagnostics platform developed by Whiteside’s group [3]. Miniaturisation obviously leads to significant reagent cost saving provided it does not run into detection-limit issues. Multiplexed detection also simplifies the analysis since manual dispensing into individual reaction tubes is not required. For example, the chip used with Gene-Z does not require external active elements for sealing, pumping, or distributing samples into individual reaction wells, eliminating potential for contamination between chips or to the device.
Type of genetic assays on POCs
So what types of genetic assays are more likely to move to POCs first? For regions with excellent centralised screening, it may be those assays where getting the results quickly using POCs saves lives or has tangible long term benefits, e.g. quickly deterring the infection and its antibiotic resistance. The leading example of this is MRSA, for which resistance has continuously increased over the past few decades. It is now known that patients are more likely to acquire MRSA in wards where the organism is screened by culturing compared to rapid molecular techniques. In such cases, detection of antibiotic resistance genes using a panel of genetic assays and POCs would minimise the practice of administering broad spectrum antibiotics because the results are not available soon enough.
In limited resource settings, the examples of genetic testing by POCs are literally endless – TB, malaria, dengue fever, HIV, flu, fungal infections and so on. This is because very little or no centralised screening occurs in such scenarios. The ability to measure dengue virus, for example, in 1–4 µl of blood could provide better tools to the 2.5 billion people who are at risk of infection and the 50–100 million people who do contract it every year. Similarly, multidrug-resistant and extensively drug-resistant TB is a global concern due to the high cost of treatment. At present, large numbers of mutations cannot be measured simultaneously using POCs. However, except the fact that isothermal mutation assays are fewer and the success rate for primer development is much lower than the signature marker probe/primer based assays, there are no technical barriers. The availability of a simple isothermal mutation assay will go a long way in making many genotyping-based diagnostics available on POCs.
In the long run, POCs may even be used to detect and quantify genetic markers associated with non-infectious diseases, such as cancer, and selected assays focusing on human genetics. Globally, cancer is responsible for 7.6 million deaths (2008 data) and projected to be rise to 13.1 million by 2030. Simple and quantitative tools capable of measuring a panel of markers may play an additional role – they may help collect data related to potentially useful but un-tested markers. Both PCR and isothermal-based assays are amenable to this application using circulating tumour cells, circulating free DNA/RNA, gene mutations, and microRNA panels. Currently utilised methods of cancer detection are highly invasive and time consuming. Minimally invasive methods on POCs may significantly increase the deployment of such capabilities.
Why do we need the wireless connectivity for POCs?
With POCs, comes the question of connectivity. Is it a must or good to have? We envision that it is important to have, but that a less useful form of device may be deployed without connectivity. Wireless connectivity via cellular phones has many advantages. Paramount among them is access to the physician and/or nurse for expert input and support. Technical advantages are automated data transfer, increased efficiency in reporting, saving time, lower equipment costs due to complexity of a touch-screen user interface and the computational power needed for data analysis.
The use of cellphones is an obvious possibility due to its ubiquitous availability and the vast network of mobile services. “There are 7 billion people on Earth; 5.1 billion own a cellphone; 4.2 billion own a toothbrush (Mobile Marketing Association Asia, 2011). By 2015 it is estimated that one third of cellphone users will have used mobile health solution in some form. However, POC genetic diagnostics and mobile networking have not yet crossed their paths. Some gene analysers (e.g. Gene-Z, Hunter) already have network enabled wireless connectivity to bridge these paths. More work is needed, however. One critical element is that transfer of data including through wireless mode must meet the requirements of the Health Insurance Portability and Accountability Act of 1996 (HIPAA) Privacy and Security Rules set by the U.S. Department of Health and Human Services. FDA clearance standards and specifications are still evolving for this area [4].
Impacts of the resulting products and devices are expected on both communicable and non-communicable diseases. Qualcomm Life provides a platform (2Net), that could be used for many different applications. According to them, “The 2Net platform is designed as an end-to-end, technology-agnostic cloud-based service that interconnects medical devices so that information is easily accessible by device users and their healthcare providers and caregivers” (http://www.qualcommlife.com/). Although the famous medical scanner or Tricorder of Star Wars fame is not yet possible, the recently announced $10 million prize by X-Prize Institute sponsored by Qualcom Life, for developing a Tricorder that can diagnose a set of 15 diseases without the intervention of the physician and weighs less than 2.3 kg is not too far from reality. In ten years, we should expect nothing less than a POC platform that is capable of sequencing-based diagnostics with assay cost of less than a dollar.
References
1. Craw P, Balachandran W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: a critical review. Lab Chip 2012; 12: 2469–2486.
2. Stedtfeld RD, Tourlousse DM, Seyrig G, Stedtfeld TM, Kronlein M, Price S, Ahmad F, Erdogan G, Tiedje JM, Hashsham SA. Gene-Z: a device for point of care genetic testing using a smartphone. Lab Chip 2012; 12: 1454–1462.
3. Martinez AW, Phillips ST, Whitesides GM, Carrilho E. Diagnostics for the developing world: microfluidic paper-based analytical devices. Anal Chem 2010; 82: 3–10.
4. Draft Guidance for Industry and Food and Drug Administration Staff – Mobile Medical Applications. July 21, 2011. www.fda.gov/medicaldevices/deviceregulationandguidance/guidancedocuments/ucm263280.htm.
The authors
Robert Stedtfeld PhD, Maggie Kronlein and
Syed Hashsham, PhD*
Civil and Environmental Engineering
1449 Engineering Research Court Rm A127
Michigan State University
East Lansing, MI 48824, USA
*Corresponding author:
E-mail: hashsham@egr.msu.edu