Prokaryotic-expressed recombinant nucleocapsid protein for the detection of coronavirus infection
The early diagnosis of common colds caused by coronavirus is a crucial step in preventing the recurrence of a global outbreak. The goals of this article are to discuss a prokaryotic-expressed recombinant nucleocapsid protein used in the development of a sensitive diagnostic assay for the diagnosis of human coronavirus infection.
by Dr Ming-Hon Hou
An overview of coronavirus
Human coronavirus (HCoV) was identified in the 1960s and has generally been associated with symptoms of the common cold. Although HCoV infections are generally mild, more severe upper and lower respiratory tract infections, such as bronchiolitis and pneumonia, which are particularly severe in infants, elderly individuals, and immunocompromised patients, have been documented. There have also been reports of clusters of HCoV infections that cause pneumonia in adults. In addition, a previous study reported that the neurotropism and neuroinvasion of HCoV are associated with multiple sclerosis.
In recent years, several emerging human coronaviruses have been discovered, and between 2003 and 2004, the SARS-CoV outbreak caused a worldwide epidemic that had a significant economic impact in countries where the disease outbreak occurred. Phylogenetic analyses have shown that SARS-CoV contains sequences that are closely related to sequences found in the betacoronaviruses. In 2004, another alphacoronavirus, HCoV-NL63, which was isolated from a 7-month-old child suffering from bronchiolitis and conjunctivitis, was reported in the Netherlands. In 2005, a novel betacoronavirus, HKU1 was found in patients with respiratory tract infections. Recently, a novel SARS-like coronavirus was found in patients with respiratory tract infections in the Middle East.
The RNA genomes of coronaviruses include genes encoding the structural proteins S (spike), M (matrix), E (envelope), and N (nucleocapsid). Additionally, some coronaviruses encode a third glycoprotein, HE (hemagglutinin-esterase), which is present in most of the betacoronaviruses. A helical nucleocapsid exists in the centre of the viral particle. The primary function of CoV N protein (NP) is to recognize a stretch of RNA that serves as a packaging signal, leading to the formation of the ribonucleoprotein (RNP) complex or to a long helical nucleocapsid structure during viral assembly. The formation of the RNP is important for maintaining the RNA in an ordered conformation suitable for replication and transcription of the viral genome. The CoV NP was shown to be involved in the regulation of cellular processes, such as gene transcription, actin reorganization, host cell cycle progression, and apoptosis.
Coronaviruses cause colds of mild to moderate severity and are transmitted by aerosols of respiratory secretions, the fecal–oral route, and mechanical transmission. The most common symptoms of coronavirus infection are nasal catarrh and a sore throat, and the illness typically lasts approximately 6 to 8 days. The early diagnosis of common colds caused by a coronavirus is an important step in preventing the recurrence of a global outbreak. Previously, rapid viral diagnosis has also been critical in the control of epidemics and the management of SARS patients. HCoVs are difficult to detect, and the current diagnosis of coronaviral infection is based on reverse transcription polymerase chain reaction with real-time PCR and antibody detection.
Previous studies have shown that NPs are the immunodominant domain in hosts infected with several coronaviruses. Additionally, it has been shown that NPs can accumulate intracellularly before being packaged into mature viruses and are the most abundant viral protein. NP is involved in the pathological reaction to human coronavirus and is a key antigen for the development of a sensitive diagnostic assay. These characteristics make NP a suitable candidate for the early diagnosis of coronavirus infection.
Nucleocapsid protein for coronavirus serodiagnosis
NP is involved in the pathological reaction to CoV infection and has been used in the development of a sensitive diagnostic assay. Previous studies reported that NP can be detected in the serum samples of SARS patients as early as 1 day after disease onset. Prokaryotic-expressed NPs have successfully been used as antigens for the detection of antibodies specific to many viruses, including SARS-CoV and several animal coronaviruses, and were produced for establishing an antigen-capture ELISA (or Western blot assay) for the diagnosis of HCoV infection
These methods are highly sensitive and specific. For example, Shi et al. [10] used recombinant SARS-CoV NP to establish an antigen-capture ELISA for SARS diagnosis. Anti-NP antibodies could be detected in approximately 90% of SARS patients 11 to 61 days after illness. No false positives were observed in non-SARS patients or health care workers.
An immunofluorescence assay is the gold standard for the detection of SARS. However, it requires efficient SARS-CoV replication in vitro to use whole virus or infected cell lysates as antigens. There are several reasons for selecting a recombinant protein rather than whole virus for this assay. The prokaryotic expression system is high yield, inexpensive, highly efficient, does not require viral cultures, and is non-toxic. Despite these advantages, viral proteins expressed in prokaryotic cells lack post-translational modifications that are present in proteins expressed in baculovirus expression systems.
Using recombinant nucleocapsid protein as an antigen for coronavirus infection diagnosis: one recent case study
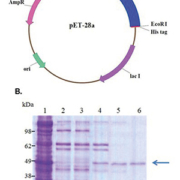
HCoV is distributed worldwide. Recently, we produced soluble recombinant human coronavirus OC43 (HCoV-OC43) NP to analyse the antigenicity of the betacoronavirus HCoV-OC43 NP. To express soluble HCoV-OC43 NP as a set of fusion proteins in E. coli, the NP gene was cloned into the pET-28a expression vector. His-tagged NP was purified from the soluble fraction using Ni-NTA column chromatography [Figure 1]. The yield from 1 L of bacterial culture was as large as 10 mg of pure NP after extraction and column chromatography. A recombinant protein-based Western blot assay was used to screen human serum from young adults, middle-aged and elderly patients with respiratory infection symptoms and cord blood units.
Western blotting is generally accepted as the most effective method for unequivocally locating linear or continuous immunodominant epitopes. Pohl-Kooppe et al. [8] also reported that Western blotting is a more sensitive test system than an immunofluorescence assay for the analysis of sera from pediatric groups. Our results showed that approximately 80–90% of serum samples from young adults and middle-aged and elderly patients with respiratory infections reacted strongly to the HCoV-OC43 NP, indicating prior exposure to this disease. In addition, the serum samples tested in this study were 81% seropositive for HCo-229E NP [Fig. 2].
This finding is consistent with previous epidemiological surveys that concluded that seroprevalence increases rapidly during childhood, attaining a seroprevalence rate of up to 90% in adults. Additionally, antibodies against HCoV-OC43 NP were detected in over 90% of cord blood samples tested. Maternally acquired antibodies may help to protect a newborn baby from HCoV-OC43 infection, although this protection appears to wane by 4 to 5 months of age. HCoV is responsible for approximately 30% of all common colds, and it is expected that 80–90% of serum samples from healthy donors and patients have antibodies to HCoV-OC43.
CoV NPs contain multiple immunodominant epitopes and antigenic sites. To compare the immunoreactivity of the three structural regions of HCoV-OC43 NP, three truncated recombinant fragments [aa 1–173 (the N-terminal domain), aa 174–300 (the central region), and aa 301–448 (the C-terminal domain)] were produced in E. coli; these regions were chosen based on PONDR (predictor of naturally disordered regions) predictions. The reactivity of human serum against these fragments was determined through Western blotting. The human serum reacted strongly with the central region and the C-terminal domain of the NP, whereas the N-terminal domain demonstrated low reactivity with the antibody. The findings of the current study are consistent with those of Chen et al. [2], who found that the antigenicity of the C-terminus of SARS-CoV NP was higher than that of the N-terminus.
The polyclonal antibody against coronavirus NP could be used to develop a rapid, easy and specific diagnostic tool for the detection of HCoV infections through immunofluorescence or ELISA-based tests. Many studies have reported that NP polyclonal antibody does not cross-react with other human CoV NPs, including those of SARS-CoV and HCoV-229E, despite the presence of highly conserved motifs in these coronavirus NPs. Previous studies also showed that the anti-SARS CoV NP and anti-HCoV-229E NP polyclonal antibodies did not cross-react with other human CoV NPs.
In our recent studies, using purified recombinant NP as an antigen, a polyclonal antibody was generated from rabbit serum with specificity for HCoV-OC43 NP; this antibody reacted specifically with HCoV-OC43 NP and did not cross-react with other human CoV NPs (including those of SARS-CoV and 229E) through Western blotting.
Conclusion
A novel SARS-like coronavirus was found in patients with respiratory tract infections in the Middle East. Thus, new and convenient diagnostic methods for CoV infection are urgently needed. The prokaryotic expression of recombinant HCoV NP is suitable for high-sensitivity, highly specific antibody production and can be used for the epidemiological screening of HCoV infection in the future.
References
1. Che XY, Qiu LW, Liao ZY, Wang YD, Wen K, Pan YX, Hao W, Mei YB, Cheng VC, Yuen KY. Antigenic cross-reactivity between severe acute respiratory syndrome-associated coronavirus and human coronaviruses 229E and OC43. J Infect Dis 2005; 191: 2033–7.
2. Chen Z, Pei D, Jiang L, Song Y, Wang J, Wang H, Zhou D, Zhai J, Du Z, Li B, Qiu M, Han Y, Guo Z, Yang R. Antigenicity analysis of different regions of the severe acute respiratory syndrome coronavirus nucleocapsid protein. Clinical Chem 2004; 50: 988–95.
3. He Q, Chong KH, Chng HH, Leung B, Ling AE, Wei T, Chan SW, Ooi EE, Kwang J. Development of a Western blot assay for detection of antibodies against coronavirus causing severe acute respiratory syndrome. Clin Diagn Lab Immunol 2004; 114: 417–22.
4. Huang CY, Hsu YL, Chiang WL, Hou MH. Elucidation of the stability and functional regions of the human coronavirus OC43 nucleocapsid protein. Protein Sci 2009; 18: 2209–18.
5. Huang LR, Chiu CM, Yeh SH, Huang WH, Hsueh PR, Yang WZ, Yang JY, Su IJ, Chang SC, Chen PJ. Evaluation of antibody responses against SARS coronaviral nucleocapsid or spike proteins by immunoblotting or ELISA. Journal Med Virol 2004; 73: 338–46.
6. Liang FY, Lin LC, Ying TH, Yao CW, Tang TK, Chen YW, Hou MH. Immunoreactivity characterisation of the three structural regions of the human coronavirus OC43 nucleocapsid protein by Western blot: Implications for the diagnosis of coronavirus infection. J Virol Methods 2013; 187: 413–20.
7. Mourez T, Vabret A, Han Y, Dina J, Legrand L, Corbet S, Freymuth F. Baculovirus expression of HCoV-OC43 nucleocapsid protein and development of a Western blot assay for detection of human antibodies against HCoV-OC43. J Virol Methods 2007; 139: 175–80.
8. Pohl-Koppe A, Raabe T, Siddell SG, ter Meulen V. Detection of human coronavirus 229E-specific antibodies using recombinant fusion proteins. J Virol Methods 1995; 55: 175–83.
9. Shao X, Guo X, Esper F, Weibel C, Kahn JS. Seroepidemiology of group I human coronaviruses in children. J Clin Virol 2007; 40: 207–13.
10. Shi Y, Yi Y, Li P, Kuang T, Li L, Dong M, Ma Q, Cao C. Diagnosis of severe acute respiratory syndrome (SARS) by detection of SARS coronavirus nucleocapsid antibodies in an antigen-capturing enzyme-linked immunosorbent assay. J Clin Microbiol 2003; 41; 5781–2.
11. Timani KA, Ye L, Zhu Y, Wu Z, Gong Z. Cloning, sequencing, expression, and purification of SARS-associated coronavirus nucleocapsid protein for serodiagnosis of SARS. J Clin Virol 2004; 30: 309–12.
The author
Ming-Hon Hou PhD
1 Biotechnology Center, National Chung Hsing University, Taichung, Taiwan
2 College of Life Science, National Chung Hsing University, Taichung, Taiwan
3 Institute of Genomics and Bioinformatics, National Chung Hsing University,
Taichung, Taiwan
E-mail: mhho@dragon.nchu.edu.tw