Proteomics as an alternative diagnostic tool for cervical cancer
Cervical cancer is mainly caused by high-risk human papillomavirus (HPV) infection. The Pap test is the gold standard for early cervical cancer diagnosis. However, the lack of Pap test accessibility accounts for the high rates of cervical cancer mostly in developing regions. Here we discuss recent proteomic approaches towards the development of novel diagnostic and prognostic putative biomarkers.
by Georgia Kontostathi, Dr Jerome Zoidakis, Prof. Nicholas P. Anagnou, Prof. Kalliopi I. Pappa and Dr Manousos Makridakis
Background
Cervical cancer is one of the most common gynecological cancers. It shares many common characteristics such as pathways and regulatory networks with vulvar and endometrial cancer [1]. The majority of cervical cancer incidents are attributed to HPV infection by high-risk oncogenic HPV types (mostly HPV16 and HPV18). HPV infects the basal membrane of cervical epithelium leading to upregulated expression of E6 and E7 oncogenes that cause specific histological lesions such as CIN1 [cervical intraepithelial neoplasia or low-grade squamous intraepithelial lesions (LSIL)], CIN2 and CIN3 [or high-grade squamous intraepithelial lesions (HSIL)] [2, 3].
Cervical cancer is the fourth most common cancer in women worldwide, regarding incidence and mortality. It was responsible for 528 000 incidents of malignancy and 266 000 deaths in 2012, of which more than 85% occurred in developing regions [4]. The high number of cervical cancer cases in developing countries is mainly attributed to the limited availability of diagnostic tools such as Pap smear tests or HPV DNA genotyping that enable detection of early-stage lesions [5].
Current diagnostic methods
The Pap test is the most popular diagnostic technique and is based on the nuclear morphology evaluation of cervical epithelial cells. This test enables the detection of possible lesions at an early stage [6]. However, there is a wide range of sensitivity (from 33.8% to 94.0%) [7, 8] that reflects the main limitation of Pap test: the high inter-observer variability.
Other diagnostic options are based on direct probes, such as Southern blot for HPV genomic analysis. This technique has a relatively low sensitivity, is time-consuming and requires large amounts of purified DNA. More sensitive methods include commercially available kits like Digene’s HC2 test (not HPV type specific), which is based on detection of viral RNA by probes. It is used for patients with minor abnormalities detected by Pap test that need further confirmation. Finally, targeted amplification methods such as PCR, are ideal for viral load quantification and genotyping with high sensitivity. However, they are prone to environmental contamination and false-positive results [9]. A recent method [approved by the US Federal Drug Administration (FDA) in 2014] is Roche’s COBAS HPV test for use in primary screening [10]. A major drawback of the above HPV-based tests is the high cost and the requirements for specialized experimental facilities.
Some protein biomarkers have been proposed for early cervical cancer screening. One of them is p16INK4a (cyclin-dependent kinase inhibitor) which is highly expressed at dysplastic epithelium. A combinatorial stain of p16INK4a and the cell proliferation marker Ki-67 has been proposed for increased diagnostic sensitivity [11]. However, lack of a scoring system for immunohistochemistry has hampered the incorporation of the these biomarkers in wide cervical cancer screening. Also, squamous cell carcinoma antigen (SCCA) is a known cancer antigen isolated from tissue, which is usually measured by immunoassays such as ELISA in serum or plasma patients, with limited specificity and sensitivity [12, 13].
Similarly, targets of the E5 HPV protein [e.g. epidermal growth factor receptor, p21 and p27 inhibitors of cyclin-dependent kinase, cyclooxygenase-2 (cox-2), vascular endothelial growth factor, and caveolin-1] have been proposed for early stage cervical cancer detection. Moreover, putative markers that have been suggested are the ProEx C immunocytochemical assay (that targets the expression of topoisomerase II protein and the minichromosome maintenance complex II protein) as well as microRNAs which are regulated by E5, E6, and E7 HPV proteins [14].
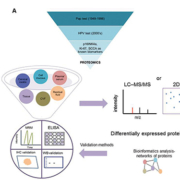
Figure 1A summarizes the different methods that have been used for the diagnosis of cervical cancer.
Current proteomic studies
The limitations of the above methods demonstrate the need for the establishment of new reliable diagnostic tests via alternative methods [15]. A novel method that can expose the association of HPV infection and cellular transformation at the molecular level is proteomics [16]. Cervical cancer models (cell cultures or tissue samples) have been studied via several proteomic methods which are either gel-based [two-dimensional gel electrophoresis (2DE)] or gel-free (liquid chromatography (LC)] in combination with mass spectrometry (MS) [17]. Proteomics can be used in order to reveal putative biomarkers for early-stage diagnosis. The role of systems biology that includes the integration of proteomics data with other available ‘OMICS’ datasets, such as genomics and transcriptomics, appears to be vital towards the direction of personalized cervical cancer medicine [18].
In this review, we present some of the most recent and interesting proteomic studies for putative biomarkers in a variety of clinical samples. Diagnostic, prognostic and predictive biomarkers have been assessed in tissue, plasma/serum, cell biopsies/cervical swabs, residual fluid from cell biopsies, cell mucous and cervicovaginal fluid (CVF) by proteomic approaches [19]. Some of the most interesting studies from each category of clinical samples are reported below. The proteomics approach workflow is presented in Figure 1B.
A proteomic study, highlighted keratin 17 as a prognostic biomarker with characteristic gradual increase from normal toward cancer stage in tissue samples. Specifically, stage specific tissue samples [normal squamous mucosa, LSIL, HSIL, and SCC (total number of samples N=22] were analysed by laser capture microdissection in combination with multidimensional liquid chromatography and tandem MS (LC-MS/MS). Proteomics analysis demonstrated differentially expressed and statistically significant proteins and after bioinformatics analysis (gene ontology) keratin 4 and keratin 17 were chosen for validation by immunohistochemistry. The gradual increase from normal towards cancer stage of keratin 17 shown by proteomics, was in total agreement with the immunohistochemistry results. Kaplan–Meier curves of keratin 17 expression and general survival of cervical cancer patients revealed a strong correlation of high keratin 17 expression with poor survival, adding prognostic value to this protein [20]. Another proteomics study focused on the pelvic lymph node metastasis (PLNM) clinical status, which is important in terms of prognosis and treatment of cervical cancer. A two-dimensional difference gel electrophoresis (2D DIGE)/matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF)-MS approach was used to compare cervical tissues of patients with PLNM (N=8) to cervical tissues from patients without lymph node metastasis (NPLNM) (N=10). Analysis led to the identification of 41 differentially expressed and statistically significant proteins. Some of them (FABP5, HspB1, and MnSOD) were validated in the PLNM group compared to the NPLNM, by Western blot and immunohistochemistry [21].
The combination of Isobaric tag for relative and absolute quantitation (iTRAQ) and targeted mass spectrometric quantification was used in order to analyse serum from CIN, early- (CES), and late-stage (CLS) cervical cancer patients. A panel of six differentially expressed proteins (alpha-1-acid glycoprotein1, alpha-1-antitrypsin, serotransferrin, haptoglobin, alpha-2-HS-glycoprotein, and vitamin D-binding protein) was validated by MRM (multiple reaction monitoring) in an independent set of 229 serum samples consisting of controls (N=49), CIN-1 to 3 designated as CIN (N=48), CES (N = 49), CLS (N=34), and ovarian cancer (N=49). The above panel discriminated patients with CIN from healthy controls with a sensitivity of 67% and specificity of 88%. Combination of the specific panel with SCCA, a well-studied putative biomarker for cervical cancer, improved discrimination of CIN, CES, and CLS patients from healthy control. Briefly, upon the comparison of the CES versus healthy group, the area under the curve was 0.86 (sensitivity/specificity = 71/90%), when using the six-protein panel and SCCA [12].
An alternative strategy was used by Boylan et al., in order to study the proteome of Pap test clinical samples. The cell-free residual fluid from cell biopsies was collected, proteins were isolated (acetone precipitation) and their concentration upon resolubilization was determined by Bradford assay. Filter aided sample preparation followed by LC-MS/MS analysis yielded 300 protein identifications per sample and 700 unique protein identifications after pooling the samples. Many of the proteins had similar biological functions to the ones identified from CVF. Thus, residual fluid could be an alternative material for the study of proteins related to cervical dysplasia [22].
The cell mucous proteome from 25 HPV-positive and pre invasive cervical disease samples has been investigated by a combination of 2DE MS, gel-based LC-MS/MS and 2DE MS after depletion of highly abundant proteins (e.g. albumin). The above approaches were combined and 107 unique proteins were identified. A bioinformatics study showed that they are related to metabolism, immune response, and transport. Proteins like acute-phase plasma proteins, α-1-antichymotrypsin and α-1-antitrypsin, were found to be both phosphorylated and glycosylated after posttranslational modifications evaluation with appropriate fluorescent dyes [23].
Table 1 highlights the above clinical proteomics studies and some of the most promising putative biomarkers that were identified.
Discussion and conclusions
Cervical cancer is the fourth most common cancer in women. The Pap test is a very efficient diagnostic approach in terms of specificity (77.8-98.8%) but has a variable sensitivity (33.8% to 94.0%) [8). Molecular tests such as HPV DNA detection by PCR and/or hybridization with adequate probes are the reference methods for the detection of HPV. The aforementioned methods are often expensive and unavailable in the developing regions where cervical cancer is very frequent (85% of cervical cancer cases). A promising idea is that proteomics will facilitate the discovery of novel biomarkers that will enable the future cervical cancer screening in developing countries in the form of antibody-based practical diagnostic self-tests (like pregnancy tests). New and advanced proteomic techniques like MRM could validate several biomarkers that will eventually be combined into panels for more accurate testing in the above diagnostic tests. Of course, the future of proteomics studies is not only promising but also challenging. New aspects of research should be taken into consideration. Redox proteomics will be used for the exploration of proteins oxidation status in order to reveal the interaction of oxidative stress and tumour development. In particular, the oxidative status of proteins in HPV-related cervical cancer cells was explored via oxidative isotope-coded affinity tags (OxICAT) and voltage-dependent anion channel 1 (VDAC1) was found to be highly oxidized in HPV-positive cervical cancer cells [24]. The important role of post-translational modifications (PTMs) such as phosphorylation and glycosylation should be thoroughly examined. The combination of systems biology and proteomics offers the possibility to elucidate cervical cancer mechanisms and identify potential biomarkers for early-stage detection.
References
1. Pappa KI, Polyzos A, Jacob-Hirsch J, Amariglio N, Vlachos GD, Loutradis D, Anagnou NP. Profiling of discrete gynecological cancers reveals novel transcriptional modules and common features shared by other cancer types and embryonic stem cells. PloS One 2015; 10(11): e0142229.
2. Doorbar J, Quint W, Banks L, Bravo IG, Stoler M, Broker TR, Stanley MA. The biology and life-cycle of human papillomaviruses. Vaccine 2012; 30(Suppl 5): F55–70.
3. Schiffman MH, Castle P. Epidemiologic studies of a necessary causal risk factor: human papillomavirus infection and cervical neoplasia. J Natl Cancer Inst. 2003; 95(6): E2.
4. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015; 136(5): E359–386.
5. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011; 61(2): 69–90.
6. Wilting SM, Smeets SJ, Snijders PJ, van Wieringen WN, van de Wiel MA, Meijer GA, Ylstra B, Leemans CR, Meijer CJ, et al. Genomic profiling identifies common HPV-associated chromosomal alterations in squamous cell carcinomas of cervix and head and neck. BMC Med Genomics 2009; 2: 32.
7. Nayar R, Wilbur DC. The Pap test and Bethesda 2014. Cancer Cytopathol. 2015; 123(5): 271–281.
8. Wright TC, Jr. HPV DNA testing for cervical cancer screening. FIGO 26th Annual Report on the Results of Treatment in Gynecological Cancer. Int J Gynaecol Obstet. 2006; 95(Suppl 1): S239–246.
9. Hubbard RA. Human papillomavirus testing methods. Arch Pathol Lab Med. 2003; 127(8): 940–945.
10. Hogarth S, Hopkins M, Rotolo D. Technological accretion in diagnostics: HPV testing and cytology in cervical cancer screening. In: Consoli D, Mina A, Nelson RR, Ramlogan R (eds) Medical Innovation: Science, Technology and Practice. Routledge 2015.
11. von Knebel Doeberitz M, Reuschenbach M, Schmidt D, Bergeron C. Biomarkers for cervical cancer screening: the role of p16(INK4a) to highlight transforming HPV infections. Expert Rev Proteomics 2012; 9(2): 149–163.
12. Boichenko AP, Govorukhina N, Klip HG, van der Zee AG, Guzel C, Luider TM, Bischoff R. A panel of regulated proteins in serum from patients with cervical intraepithelial neoplasia and cervical cancer. J Proteome Res. 2014; 13(11): 4995–5007.
13. Kato H, Torigoe T. Radioimmunoassay for tumor antigen of human cervical squamous cell carcinoma. Cancer 1977; 40(4): 1621–1628.
14. de Freitas AC, Coimbra EC, Leitao Mda C. Molecular targets of HPV oncoproteins: potential biomarkers for cervical carcinogenesis. Biochim Biophys Acta 2014; 1845(2): 91–103.
15. Wentzensen N, von Knebel Doeberitz M. Biomarkers in cervical cancer screening. Dis Markers 2007; 23(4): 315–330.
16. Lomnytska M, Souchelnytskyi S. Markers of breast and gynecological malignancies: the clinical approach of proteomics-based studies. Proteomics Clin appl. 2007; 1(9): 1090–1101.
17. Di Domenico F, De Marco F, Perluigi M. Proteomics strategies to analyze HPV-transformed cells: relevance to cervical cancer. Expert Rev Proteomics 2013; 10(5): 461–472.
18. Breuer EK, Murph MM. The role of proteomics in the diagnosis and treatment of women’s cancers: current trends in technology and future opportunities. Int J Proteomics 2011; 2011: pii: 373584.
19. Kontostathi G, Zoidakis J, Anagnou NP, Pappa KI, Vlahou A, Makridakis M. Proteomics approaches in cervical cancer: focus on the discovery of biomarkers for diagnosis and drug treatment monitoring. Exp Rev Proteomics 2016; 13(8): 731–745.
20. Escobar-Hoyos LF, Yang J, Zhu J, Cavallo JA, Zhai H, Burke S, Koller A, Chen EI, Shroyer KR. Keratin 17 in premalignant and malignant squamous lesions of the cervix: proteomic discovery and immunohistochemical validation as a diagnostic and prognostic biomarker. Mod Pathol. 2014; 27(4): 621–630.
21. Wang W, Jia HL, Huang JM, Liang YC, Tan H, Geng HZ, Guo LY, Yao SZ. Identification of biomarkers for lymph node metastasis in early-stage cervical cancer by tissue-based proteomics. Br J Cancer 2014; 110(7): 1748–1758.
22. Boylan KL, Afiuni-Zadeh S, Geller MA, Hickey K, Griffin TJ, Pambuccian SE, Skubitz AP. A feasibility study to identify proteins in the residual Pap test fluid of women with normal cytology by mass spectrometry-based proteomics. Clin Proteomics 2014; 11(1): 30.
23. Panicker G, Ye Y, Wang D, Unger ER. Characterization of the human cervical mucous proteome. Clini Proteomics 2010; 6(1–2): 18–28.
24. Zhang C, Ding W, Liu Y, Hu Z, Zhu D, Wang X, Yu L, Wang L, Shen H, et al. Proteomics-based identification of VDAC1 as a tumor promoter in cervical carcinoma. Oncotarget 2016; doi: 10.18632/oncotarget.10562 [Epub ahead of print].
The authors
Georgia Kontostathi1,2 MSc; Jerome Zoidakis1 PhD; Nicholas P. Anagnou2,3 MD, PhD; Kalliopi I. Pappa3,4 MD, PhD; Manousos Makridakis*1 PhD
1Biotechnology Division, Biomedical Research Foundation, Academy of Athens (BRFAA), Athens, Greece
2Laboratory of Biology, University of
Athens School of Medicine, Athens, Greece
3Cell and Gene Therapy Laboratory,
Biomedical Research Foundation,
Academy of Athens (BRFAA), Athens, Greece
4First Department of Obstetrics and
Gynecology, University of Athens School of Medicine, Athens, Greece
*Corresponding author
E-mail: mmakrid@bioacademy.gr