Using whole-genome sequencing to predict antimicrobial resistance
Thousands of strain-specific whole-genome sequences are now available for a wide range of pathogenic bacteria. Using these data, approaches based on machine learning can now be used to predict the results of antimicrobial susceptibility tests from sequence alone. Recent studies have demonstrated the ability to predict minimum inhibitory concentrations with accuracies up to 95 %. Employing these tools to prioritize antibiotic treatment could improve patient outcomes and help to avoid the antibiotic resistance crisis.
by Dr Jonathan M. Monk
Importance of antimicrobial resistance (AMR) prediction
Today over 700 000 people die of antibiotic resistant infections per year [1]. Frighteningly, it has been estimated that this number could rise to 10 million deaths per year if nothing is done to stop the increase and spread of antibiotic resistant bacteria [2]. To help combat this threat it is critical to limit the use of ineffective antibiotics and to prescribe the appropriate antimicrobial therapy to patients as quickly as possible. Although antimicrobial susceptibility testing is now routine in microbiology laboratories, this testing often takes too long to impact clinical diagnosis.
New tools that rapidly predict antibiotic resistance could improve antibiotic stewardship and, when effectively implemented, have led to reductions in levels of resistant bacteria in hospitals [3]. Thus, accurately diagnosing antibiotic resistant bacteria would avoid the evolutionary pressures that accelerate resistance and would aid antibiotic stewardship approaches. This could enable physicians to select the optimal antibiotic regimen to cure a patient, rather than enhancing a given strain’s resistance. Whole-genome sequencing may offer this possibility.
The genomics revolution has made available thousands of strain-specific whole-genome sequences (WGS) for a range of pathogenic bacteria. For example the Pathosystems Resource Integration Center (PATRIC) [the all-bacterial Bioinformatics Resource Center (BRC) funded by the National Institute of Allergy and Infectious Diseases (NIAID)] currently contains over 15 000 Escherichia genomes, more than 14 000 Staphylococcus genomes and nearly 11 000 Mycobacteria genomes [4]. Increasingly, these genomes are coupled with clinical metadata, including minimum inhibitory concentration (MIC) values for various antibiotics.
This large-scale coupling of resistance data with strain-specific genome sequences enables machine learning and other big-data science approaches to study and predict antibiotic resistance. For example, it is now possible to apply case-control studies whereby a group of strains that exhibit a biological phenotype (e.g. antibiotic resistance) is compared to a group of strains that do not. Machine learning techniques can be used to identify biomarkers (e.g. presence/absence of genes or mutations) that are predictive of a given phenotype. These biomarkers can then serve as a basis for diagnostic tests.
Here we discuss recent literature using machine learning approaches to predict antibiotic resistance and highlight considerations required for their application.
Introduction to machine learning approaches for WGS-based prediction of AMR
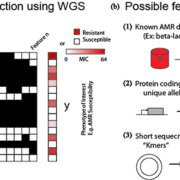
Setting up a machine learning problem involves breaking data into two groups (Fig. 1a):
(1) The y-array containing genomes or samples (m rows) matched with the phenotype to be predicted. In the case of AMR prediction, a phenotype could be binary e.g. ‘resistant’ versus ‘susceptible’ or the actual experimentally measured MIC. Predicting MICs is often preferable owing to changing breakpoints used to define resistance. For example, a given strain with a MIC of 8 µg/mL gentamicin may have previously been classified as resistant, but new CLSI 2017 guidelines specify that gentamicin resistance requires a MIC above 16 µg/mL. This can lead to inconsistent AMR annotations that can confound binary predictions.
(2) The X-matrix containing the samples (m rows) and their associated features (n columns) that will be used to make a prediction. Features range from those that are completely knowledge-based, such as the presence of genes known to confer antibiotic resistance (e.g. a beta-lactamase) to those that require no previous knowledge such as the presence of short (~10 bp) segments of DNA on the chromosome (Fig. 1b). These features have unique benefits and drawbacks that have been used in several recent studies described below.
Selecting appropriate features for AMR prediction
Knowledge-based features
Knowledge-based features can be obtained by mapping a genome of interest using curated databases of gene products that have already been demonstrated to confer antibiotic resistance. As of February 2019, the Comprehensive Antibiotic Resistance Database (CARD) houses 2553 reference sequences and 1216 SNPs demonstrated to confer resistances for 79 different pathogens [5]. These approaches are akin to laboratory tools that offer PCR-based identification of AMR determinants, such as BioFire. Models trained using previously annotated features often have good accuracies and are easier to interpret because of the accumulated knowledge present in such predictions [6, 7].
However, despite their high accuracy, these tools are limited because they often require many rounds of multiple sequence alignment, which can become computationally expensive at large scale. Also, reliance on known AMR determinants may cause such algorithms to miss newly evolved resistance mechanisms. A useful machine learning approach should be capable of analysing future outbreaks and identifying new mechanisms of resistance, rather than being limited to past knowledge.
Gene- and allele-based features
An approach that balances these two extremes involves assembling features by annotating the genome for known protein coding genes, but keeping the feature types agnostic, for example by including genes with functions ranging from cell replication, to cell wall synthesis to metabolism. This approach has the advantage of not requiring known determinants of antimicrobial resistance but does still require annotated genomes, potentially biasing results by annotation methods.
Recent studies have used this approach to predict antibiotic resistance in E. coli with accuracies above 90 % [8]. Importantly, this approach identified features that outperformed genes established in the literature. Such an approach can go even deeper by breaking the genes down into their constituent alleles to account for potential mutations in each coding sequence. Another study took this approach to examine 1595 strains of M. tuberculosis and identified 33 known AMR-conferring genes and 24 new potentially novel antibiotic resistance conferring genes [9]. Thus, methods that rely on several features extracted from the genome, rather than restricting them to those with previous knowledge, can be used to accurately predict AMR and identify novel mechanisms of resistance making them extensible to mutations and mechanisms of resistance that may emerge in the future.
Kmer feature selection
A contrasting approach that can identify new mechanisms of resistance and requires no a priori knowledge involves breaking up a genome into short (~10 bp) long segments of DNA and using these to create ‘features’ from short segments of DNA on the genome, known as ‘kmers’. All genomes in the collection can be divided into kmers that are then added to the X-matrix where presence of a specific kmer becomes a feature. Thus, this kmer-based approach contrasts with knowledge-based methods to predict AMR that rely on a database of curated genes and mutations previously shown to confer antibiotic resistance.
Studies of A. baumannii, S. aureus, S. pneumoniae, K. pneumoniae and collections of over 5000 Salmonella genomes have demonstrated ability to predict MIC with an average accuracies above 90 % within +/−1 twofold dilution step [10–12]. Unfortunately, this high accuracy and ability to predict new mechanisms of resistance has the trade-off of being difficult to interpret. For example, a model may imply a strong relationship between predicted resistance and segments of the genome without annotated functions or to biological processes.
Building and evaluating a machine learning model
Once the features for a model have been selected it is time to apply a machine learning algorithm to the data. Several such algorithms exist, each with benefits and drawbacks related to accuracy and interpretability [13]. Unfortunately, often the more accurate models are difficult to interpret whereas more intelligible models have worse predictive capabilities. In healthcare applications it is vital for the treating physician to be able to understand, validate and trust a model, and thus relying on easier to interpret methods like a decision tree or simple logistic regressors may be best.
When evaluating a machine learning model it is imperative to question how the model was trained. A major pitfall for machine learning approaches is their tendency to ‘overfit’ datasets. For example, a model trained on data used to make a prediction could simply ‘remember’ that data and use it to correctly predict any point in the same training set. However, if the model is too rigid it may perform poorly on new data. Robust machine learning models avoid such overfitting by splitting the data into non-overlapping sets where ~80 % of the data is used for training and ~20 % of the data is used for tests (Fig. 2a). This splitting process should be random and be performed several times to assess the overall accuracy and sensitivity of a model, thereby limiting overfitting and ensuring that predictions remained generalizable and robust.
Once a model’s ability to predict new data is established it is finally possible to evaluate the model’s predictive performance (Fig. 2b). Thus far, we have described previous studies in terms of correct predictions and accuracies. However, it’s often more important to evaluate cases where a model fails. Requirements for AMR diagnostic devices are strict. Devices typically describe their utility in terms of error rate. Major errors (MEs) occur when susceptible genomes are incorrectly predicted to have resistant MICs. The opposite case, when resistant genomes are incorrectly assigned susceptible MICs, are termed very major errors (VMEs). US Food and Drug Administration (FDA) standards for automated systems recommend a ME rate ≤3 %. A recent study of over 5000 Salmonella genomes used kmers to train a model that demonstrated MIC predictions for 15 antibiotics with ME rates in this range [10]. The FDA standards for VME rates indicate that the lower 95 % confidence limit should be ≤1.5 % and upper limit should be ≤7.5 %. Models for seven of the 15 antibiotics in the same study had acceptable VME rates based on this requirement. Thus, such an approach would make acceptable predictions for diagnostic applications.
Summary and outlook
In summary, options for WGS-based predictions of antimicrobial susceptibility testing are becoming a reality. This brief summary limits the scope to tools and methods to predict antibiotic susceptibility from WGS. However, in the future it may be possible to combine genomic features with information from the patient, like age, gender, comorbidities, etc. Furthermore, rather than predicting only antibiotic susceptibility it would be possible to train an algorithm to predict patient outcome and adjust treatment regimens to improve patient care [14].
Such approaches are sorely needed because despite improvements in antibiotic use, the Centres for Disease Control and Prevention (CDC) estimates that approximately 50 % of antibiotics are still prescribed unnecessarily in the US at a yearly cost of $1.1 billion [15], and the annual impact of resistant infections in the US is estimated to be $20 billion in excess healthcare costs and 8 million additional days patients stay in the hospital [16]. Significant improvements in patient outcome have been observed when reducing the time of treatment with optimal antibiotic therapy [17, 18]. Rapid identification and targeted treatment of pathogenic bacteria using tools assisted by algorithms presented here would enable precision medicine for pathogens that would lower the incidence of antibiotic resistance, improve patient health, and lead to decreased hospital costs.
Figure 1. How to set up a whole-genome sequence (WGS)-based machine learning problem for antimicrobial resistance (AMR) prediction. (a) Samples (m=rows) with sequenced genomes and known phenotypes of interest [‘susceptible’ vs ‘resistant’ phenotypes or minimum inhibitory concentration (MIC) value] are used to train a machine learning model. All values to be predicted are placed into the ‘y’ array. The ‘features’ used to train a model form the columns of the X-matrix. (b) For WGS-based antimicrobial-susceptibility-test prediction possible feature types include: (1) known antibiotic resistance conferring genes or mutations, (2) annotated protein coding genes (independent of known functions) and even (3) the presence of short fragments of DNA sequence on the chromosome known as ‘kmers’. These different feature types have a trade-off between ease of interpretation (easiest for previously identified features) and ability to detect novel AMR determinants (best for short sequence fragments).
Figure 2. Evaluating predictions from a machine learning model. (a) A machine learning model that is ‘overfit’ is inflexible to new data. To ensure a model is robust enough to predict new samples, all models should be cross-validated. This process involves randomly splitting the whole dataset into training (± 80 % of samples) and testing (± 20 %) sets. The sets should be shuffled multiple times to check model accuracy across different samples and features. (b) The results of running the model on testing sets can then be compared for each randomly sampled set (different colored lines). The model’s performance is compared by calculating the area under the curve (AUC) on a plot of the true positive rate vs the false positive rate often called a receiver operating characteristic (ROC) curve. Model accuracy can be calculated from the number of true positive (TP) [model predictions, resistant (R); experimental result, R] and true negative (TN) predictions divided by the total number of predictions. However, it is often more important to gauge how a model fails: for example a false positive ‘major error’ [model prediction, R; experimental result, susceptible (S)] may lead to incorrectly withholding an effective antibiotic. Even worse, false negative ‘very major error’ predictions (model prediction, S; experimental result, R) could lead to prescribing an antibiotic that is ineffective.
References
1. The dangers of hubris on human health. Global Risks – Reports. World Economic Forum 2013 (http://reports.weforum.org/global-risks-2013/risk-case-1/the-dangers-of-hubris-on-human-health/).
2. O’Neill J. Antimicrobial resistance: tackling a crisis for the health and wealth of nations. Rev Antimicrob Resist 2014; 20: 1–16.
3. Carling P, Fung T, Killion A, Terrin N, Barza M. Favorable impact of a multidisciplinary antibiotic management program conducted during 7 years. Infect Control Hosp Epidemiol 2003; 24(9): 699–706.
4. Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, Gillespie JJ, Gough R, Hix D, Kenyon R, Machi D, Mao C, Nordberg EK, Olson R, Overbeek R, Pusch GD, Shukla M, Schulman J, Stevens RL, Sullivan DE, Vonstein V, Warren A, Will R, Wilson MJ, Yoo HS, Zhang C, Zhang Y, Sobral BW. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res 2014; 42(Database issue): D581–591.
5. Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, Doshi S, Courtot M, Lo R, Williams LE, Frye JG, Elsayegh T, Sardar D, Westman EL, Pawlowski AC, Johnson TA, Brinkman FS, Wright GD, McArthur AG. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 2017; 45(D1): D566–573.
6. Jeukens J, Freschi L, Kukavica-Ibrulj I, Emond-Rheault J-G, Tucker NP, Levesque RC. Genomics of antibiotic-resistance prediction in Pseudomonas aeruginosa. Ann N Y Acad Sci 2019; 1435(1): 5–17 (First published 2017 Online: https://nyaspubs.onlinelibrary.wiley.com/doi/abs/10.1111/nyas.13358).
7. Bradley P, Gordon NC, Walker TM, Dunn L, Heys S, Huang B, Earle S, Pankhurst LJ, Anson L, de Cesare M, Piazza P, Votintseva AA, Golubchik T, Wilson DJ, Wyllie DH, Diel R, Niemann S, Feuerriegel S, Kohl TA, Ismail N, Omar SV, Smith EG, Buck D, McVean G, et al. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat Commun 2015; 6: 10063.
8. Her H-L, Wu Y-W. A pan-genome-based machine learning approach for predicting antimicrobial resistance activities of the Escherichia coli strains. Bioinformatics 2018; 34(13): i89–i95.
9. Kavvas ES, Catoiu E, Mih N, Yurkovich JT, Seif Y, Dillon N, Heckmann D, Anand A, Yang L, Nizet V, Monk JM, Palsson BO. Machine learning and structural analysis of Mycobacterium tuberculosis pan-genome identifies genetic signatures of antibiotic resistance. Nat Commun 2018; 9(1): 4306.
10. Nguyen M, Long SW, McDermott PF, Olsen RJ, Olson R, Stevens RL, Tyson GH, Zhao S, Davis JJ. Using machine learning to predict antimicrobial MICs and associated genomic features for nontyphoidal Salmonella. J Clin Microbiol 2019; 57(2): pii: e01260-18 (http://dx.doi.org/10.1128/JCM.01260-18).
11. Davis JJ, Boisvert S, Brettin T, Kenyon RW, Mao C, Olson R, Overbeek R, Santerre J, Shukla M, Wattam AR, Will R, Xia F, Stevens R. Antimicrobial resistance prediction in PATRIC and RAST. Sci Rep 2016; 6: 27930.
12. Nguyen M, Brettin T, Long SW, Musser JM, Olsen RJ, Olson R, Shukla M, Stevens RL, Xia F, Yoo H, Davis JJ. Developing an in silico minimum inhibitory concentration panel test for Klebsiella pneumoniae. Sci Rep 2018; 8(1): 421.
13. Deo RC. Machine learning in medicine. Circulation 2015; 132(20): 1920–1930.
14. Kachroo P, Eraso JM, Beres SB, Olsen RJ, Zhu L, Nasser W, Bernard PE, Cantu CC, Saavedra MO, Arredondo MJ, Strope B, Do H, Kumaraswami M, Vuopio J, Gröndahl-Yli-Hannuksela K, Kristinsson KG, Gottfredsson M, Pesonen M, Pensar J, Davenport ER, Clark AG, Corander J, Caugant DA, Gaini S. Integrated analysis of population genomics, transcriptomics and virulence provides novel insights into Streptococcus pyogenes pathogenesis. Nat Genet 2019; 51(3): 548–559 (http://dx.doi.org/10.1038/s41588-018-0343-1).
15. Antibiotic resistance threats in the United States, 2013. US Centers for Disease Control and Prevention 2013 (https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf).
16. Fair RJ, Tor Y. Antibiotics and bacterial resistance in the 21st century. Perspect Medicin Chem 2014; 6: 25–64.
17. Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, Suppes R, Feinstein D, Zanotti S, Taiberg L, Gurka D, Kumar A, Cheang M. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 2006; 34(6): 1589–1596.
18. Palmer HR, Palavecino EL, Johnson JW, Ohl CA, Williamson JC. Clinical and microbiological implications of time-to-positivity of blood cultures in patients with Gram-negative bacilli bacteremia. Eur J Clin Microbiol Infect Dis 2013; 32(7): 955–959.
The author
Jonathan M. Monk PhD
Department of Bioengineering, UC San Diego, San Diego, California, USA
E-mail: jmonk@ucsd.edu