Liquid biopsy for diagnostic epidermal growth factor receptor gene testing in non-small cell lung cancer
Advances in circulating biomarker research have led to the use of blood samples to characterize cancer patients’ tumour DNA where a lack of tumour tissue prevents molecular testing. This is critical for non-small cell lung cancer (NSCLC) patients who require tumour molecular characterization in order to access life-extending treatments that would be denied without biopsy. Here we describe a new liquid biopsy diagnostic service for NSCLC patients at the All Wales Medical Genetics Service, Cardiff, UK.
by Dr Angharad Williams, Dr Daniel Nelmes, Helen Roberts and Dr Rachel Butler
Liquid biopsies and cell-free circulating tumour DNA (ctDNA) in clinical practice
The term ‘liquid biopsy’ comes from the sampling of a cancer patient’s tumour DNA from a simple, non-invasive blood test rather than an invasive surgical biopsy. This circulating tumour DNA (ctDNA) is a small fraction of the total cell-free circulating DNA (cfDNA) and consists of short strands of DNA shed by degrading tumour cells directly into a patient’s bloodstream. The levels of ctDNA present will vary greatly based on clinical factors such as proximity of sampling to chemotherapy or radiotherapy, as well as the burden and activity of the tumour [1].
Genetic mutations within the patient’s tumour are detectable at extremely low levels in the ctDNA in the blood [2]. The detection of such mutations provides many potential uses for ctDNA as a biomarker in disease diagnosis and screening, monitoring of therapy response and resistance and detection of minimal residual disease and relapse [3–6].
The many advantages for using ctDNA as a biomarker rest on the fact that ctDNA can be simply extracted from blood; therefore, invasive biopsy procedures can be avoided. Such simple blood sampling is beneficial if the patient is too ill for invasive surgery and is also useful if biopsy-based tumour analysis has failed; thus, unnecessary re-biopsies can be averted. Another benefit of the use of ctDNA over biopsies is that serial blood samples can be taken to replace the need for a re-biopsy to monitor a patient’s response to therapy in ‘real-time’ in the clinic. Practically, blood samples can be arranged, taken and sent for processing at a much faster pace than surgery, gaining valuable time for patients who are in need of urgent cancer-related treatments.
There are, however, potential pitfalls in using ctDNA as a diagnostic biomarker that should be considered prior to setting up a ctDNA-based diagnostic service as well as when interpreting genetic results from ctDNA (summarized in Figure 1). The greatest concern is the fragile nature of cfDNA molecules [7], which means that cfDNA will degrade in a blood sample to undetectable levels the longer that the blood is left unprocessed. Efficient centrifugation and separation of the blood to plasma and storage at −80 °C can be used to halt degradation of cfDNA. In cases where analysis of the cfDNA sample identifies no genetic mutations, this raises the important question of whether the patient was actually shedding ctDNA at the time of the blood sampling or did the ctDNA degrade prior to sample processing? This indicates the unfortunate possibility of false negative results when using ctDNA in the diagnostic setting. Another important factor to consider is that the level of a mutation in the ctDNA, which can quite often be as low as ≤1% mutated ctDNA to wild-type patient cfDNA [8]. Thus, only highly sensitive molecular analysis options should be considered for diagnostic testing strategies using ctDNA.
Molecular analysis of the epidermal growth factor receptor gene in non-small cell lung cancer patients
The epidermal growth factor receptor gene (EGFR) encodes the EGRF protein, a signalling protein that is part of the cellular pathways that control normal cell growth, differentiation and angiogenesis [9]. Approximately 10–20% of ethnically Caucasian non-small cell lung cancer (NSCLC) patients with the adenocarcinoma histological subtype will have a DNA mutation in the EGFR gene, which will activate abnormal constitutive signalling and tumorigenesis [10].
The most common sensitizing EGFR mutations, which represent 85% of known activating EGFR mutations in NSCLC, are the exon 21 point mutation c.2573T>G (p.Leu858Arg) and in-frame deletions in exon 19 [9]. These activating mutations provide a convenient target for first and second generation tyrosine kinase inhibitor (TKI) treatments such as gefitinib (Iressa®, AstraZeneca) [11–13] and act as positive predictive biomarkers for response to these drugs. Traditionally, for patients to access these TKI treatments, tumour biopsy in the form of a formalin-fixed sample is tested for evidence of these activating EGFR mutations at clinical genetic testing centres, such as the All Wales Medical Genetics Service (AWMGS) in Cardiff. However, preservation of the tumour biopsy as formalin-fixed paraffin-embedded (FFPE) tissue leads to a number of issues with genetic analysis including poor quality and yields of DNA (noted in Figure 1). Additionally, a large proportion of NSCLC patients are not well enough to have a biopsy taken and so genetic analysis of tumour DNA and subsequent access to TKI treatments is not possible. This inequity in service provision indicated a clinical need to expand current testing options for NSCLC patients to reach those patients who cannot access TKI-based stratified medicine treatment options. To address this clinical need, a ctDNA-based diagnostic NHS service was developed within AWMGS to detect activating EGFR mutations from patient blood samples in order to alleviate the need for biopsy.
In addition to the availability of first and second generation TKIs, a new third generation TKI, osimertinib (Tagrisso®, AstraZeneca), has recently been made available to a specific group of NSCLC patients. Approximately 50% of patients on first and second generation TKIs will develop an EGFR resistance mutation, c.2369C>T (p.Thr790Met) (commonly known as T790M), leading to disease progression [6]. Since October 2016, osimertinib (Tagrisso®, AstraZeneca), has been available to UK patients shown to harbour the T790M mutant in their tumour via either biopsy or ctDNA analysis through the NHS Cancer Drugs Fund [14]. CtDNA testing has become a popular method of testing for resistance mutations as it mitigates the need for a second invasive biopsy for the patient and, also, serial blood samples can be used to track the patient’s response over a period of time [15].
Establishing the ctDNA-based NSCLC stratified medicine service in the All Wales Medical Genetics Service
Since 2009, the AWMGS has been providing stratified medicine services for NSCLC patients, as well as metastatic colorectal cancer patients, melanoma and gastrointestinal stromal tumour patients in Wales. Though all of these services are based on FFPE tumour analysis, we have developed a wealth of experience in using ctDNA from blood in the field of clinical trials. By 2015, following a number of successful ctDNA-based feasibility studies by laboratory staff and research students, we were confident that we had the knowledge and expertise to bring ctDNA into service, and were one of the first laboratories in the UK to do so.
Owing to the inherent shortcomings of using ctDNA as a biomarker, discussed previously, the following questions were deliberated during validation to find the most appropriate testing methods for the diagnostic service:
- How do we best protect ctDNA in blood during sampling and shipping to the lab, to ensure that the sample that reaches us is faithful to the patient’s real mutation status?
- As important mutations in ctDNA can be at very low levels, how do we ensure we can detect a low enough range of mutations to be clinically relevant to interpretation?
To guarantee sample quality and maintain sufficient levels of ctDNA, we have imposed stringent quality measures on the blood collection and dispatch. The main requirement is that blood samples should be taken in a specialist preservative tube such as CellSave Preservative Tubes (Janssen Diagnostics) or Cell-Free DNA BCT® (Streck) and must reach the laboratory for processing within a strict 96-hour window. This was decided on after discussion with other research groups and internal investigations on the stability of ctDNA in blood and the use of preservative tubes [7].
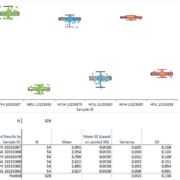
The sensitivity of the molecular ctDNA assay was paramount in our decision to use the recently developed technology droplet digital polymerase chain reaction (ddPCR) by Bio-Rad (Bio-Rad Laboratories, Inc, California, USA). ddPCR is a highly sensitive fluorescence-based PCR method with an extreme lower limit of detection of 0.0001% of mutant DNA in a wild-type background, which makes it the superior choice over other technologies such as next-generation sequencing and quantitative PCR (qPCR). Practically, in the service, we have detected EGFR mutations in patient ctDNA at an abundance as low as 0.7%.
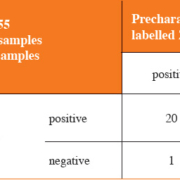
The AWMGS now provides ctDNA-based testing of the EGFR gene in NSCLC patients at both first-line testing for sensitizing mutations and for resistance mutation testing on patient progression on TKIs (Figure 2). The service launched across Wales in April 2016 and has since been expanded to provide testing for certain centres in the South West of England with funding from AstraZeneca. A year on, over 100 patients have been tested, the majority (approximately 60%) of patient referrals have been for T790M progression testing to avoid repeat biopsies for patients. Six patients, for whom TKIs were previously inaccessible due to failed FFPE-based testing or inability to biopsy, were successfully tested through the ctDNA service and are now receiving first-line EGFR TKI therapy following the detection of activating EGFR mutations in ctDNA.
Ongoing and future developments
The field of liquid biopsies is steadily gaining pace in the UK and abroad with a number of centres now providing EGFR ctDNA testing. New circulating biomarkers, such as exosomes and circulating tumour cells, are coming through from translation research and have vast potential in the field of stratified medicine. At the AWMGS, we aim to expand our current liquid biopsy testing in the near future with targets for both metastatic colorectal cancer and metastatic melanoma.
References
1. Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011; 11: 426–437.
2. Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014; 6: 224ra24.
3. Chen KZ, Lou F, Yang F, Zhang JB, Ye H, Chen W, Guan T, Zhao MY, Su XX, et al. Circulating tumor DNA detection in early-stage non-small cell lung cancer patients by targeted sequencing. Sci Rep 2016; 6: 31985.
4. Dawson S-J, Tsui DWY, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013; 368: 1199–1209.
5. Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, Dawson SJ, Piskorz AM, Jimenez-Linan M, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 2012; 4: 136ra68.
6. Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, Parkinson C, Chin SF, Kingsbury Z, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013; 497: 108–112.
7. Rothwell DG, Smith N, Morris D, Leong HS, Li Y, Hollebecque A, Ayub M, Carter L, Antonello J, et al. Genetic profiling of tumours using both circulating free DNA and circulating tumour cells isolated from the same preserved whole blood sample. Mol Oncol 2016; 10: 566–574.
8. Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem 2015; 61: 112–123.
9. Jänne PA, Engelman JA, and Johnson BE. Epidermal growth factor receptor mutations in non–small-cell lung cancer: implications for treatment and tumor biology. J Clin Onc 2005; 23: 3227–3234.
10. Li T, Kung H-J, Mack PC, Gandara DR. Genotyping and genomic profiling of non-small-cell lung cancer: implications for current and future therapies. J Clin Onc 2013; 31: 1039–1049.
11. Douillard JY, Ostoros G, Cobo M, Ciuleanu T, Cole R, McWalter G, Walker J, Dearden S, Webster A, et al. Gefitinib treatment in EGFR mutated Caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol 2014; 9: 1345–1353.
12. Goto K, Ichinose Y, Ohe Y, Yamamoto N, Negoro S, Nishio K, Itoh Y, Jiang H, Duffield E, et al. Epidermal growth factor receptor mutation status in circulating free DNA in serum: from IPASS, a phase III study of gefitinib or carboplatin/paclitaxel in non-small cell lung cancer. J Thorac Oncol 2012; 7: 115–121.
13. National Institute for Health and Care Excellence (NICE). Gefitinib for the first-line treatment of locally advanced or metastatic non-small-cell lung cancer. Technology appraisal guidance [TA192] 2010. [https: //www.nice.org.uk/guidance/ta192]
14. NICE. Osimertinib for treating locally advanced or metastatic EGFR T790M mutation-positive non-small-cell lung cancer. Final appraisal determination [TA10022] 2016. [https: //www.nice.org.uk/guidance/GID-TA10022/documents/final-appraisal-determination-document]
15. Sundaresan TK, Sequist LV, Heymach JV, Riely GJ, Jänne PA, Koch WH, Sullivan JP, Fox DB, Maher R, et al. Detection of T790M, the acquired resistance EGFR mutation, by tumor biopsy versus noninvasive blood-based analyses. Clin Cancer Res 2016; 22: 1103–1110.
The authors
Angharad Williams1 PhD, Daniel Nelmes2,3 PhD, Helen Roberts1 BSc and Rachel Butler*1 FRCPath
1All Wales Medical Genetics Service,
NHS Wales, The Institute of Medical
Genetics, Cardiff and Vale University LHB,
University Hospital of Wales, Cardiff
CF14 4XW, Wales, UK
2School of Medicine, Cardiff University, Cardiff CF14 4XN, Wales, UK
3Velindre Cancer Centre, Cardiff
CF14 2TL, Wales, UK
*Corresponding author
E-mail: Rachel.Butler@wales.nhs.uk