Risk factors for development of breast cancer bone metastasis
Breast cancer bone metastasis results in a significant reduction in patient quality of life and upon metastatic spread the disease is considered incurable. Molecules have been identified which predict the risk of developing bone metastases. This review discusses these key molecules and their potential utility within patient treatment decisions.
by Dr Steven L. Wood and Prof. Janet E. Brown
Introduction
Invasive breast cancer is diagnosed in over 55 000 women every year within the UK [1]. Despite recent advances in breast cancer treatment around 10 000 women die from breast cancer in the UK annually, almost all as a result of metastatic spread, which can occur years after apparently successful initial treatment. Over 70% of all advanced breast cancer patients develop metastatic spread to the skeleton [2, 3]. Disseminated tumour cells within bone can remain dormant for many years before finally becoming reactivated, leading primarily to bone resorption (osteolytic lesions), but also to unbalanced bone formation in response (osteoblastic lesions). Current treatments to reduce/prevent the skeletal complications in patients with established breast cancer bone metastasis (BCBM) involve the use of antiresorptive agents such as bisphosphonates [such as zoledronic acid (ZA)] [4]. An antiresorptive treatment has also been developed which utilizes antibodies directed towards key molecules within BCBM-induced bone destruction, i.e. denosumab [5]. These antiresorptive agents have been highly effective in improving quality of life for patients with BCBM, but do not improve survival once metastasis is established.
Recently, however, large studies have shown that bisphosphonates given as adjuvant treatment in early breast cancer, alongside other standard treatments, lead to a reduction in the numbers of postmenopausal patients developing bone metastasis [6]. Adjuvant treatment also leads to improved overall survival and adjuvant bisphosphonate therapy is now entering standard practice. However, these treatments are not without side effects, including osteonecrosis of the jaw [7, 8]. Since only a minority of women will develop bone metastasis, biomarkers are required to identify those patients at highest risk, enabling therapy to be targeted to those who will benefit, sparing those who will not.
Risk factors
Clinicopathological and demographic risk factors
Breast cancer is a heterogeneous disease and pathological staging and grading systems are widely used in routine practice. Although not generally specific for indicating risk of bone metastasis, these systems do categorize patients into sub-groups that determine appropriate treatment and risk of progression. The human epidermal growth factor receptor 2 (HER2) and estrogen receptor (ER) have both prognostic and predictive value and are routinely measured. ER is a hormone-regulated nuclear transcription factor that binds estrogen, with consequent expression of genes including the progesterone receptor (PR). Patients with HER2-positive breast cancer have a poorer prognosis, but targeted treatments are now available. Like ER, HER2 is also a predictive marker, identifying patients who are likely to respond to targeted treatments.
Histological subtype, tumour grade, lymph node involvement and body-mass index all impact on the general risk of metastasis and, therefore, of BCBM. It is well-recognized that bone metastases more commonly develop in ER-positive patients; they can also occur in ER-negative patients. Although these pathological categories are routinely examined, there has been a recent strong research emphasis upon the discovery of molecular risk factors for development of metastasis, including BCBM.
Molecular risk factors for bone metastasis
Genetic risk factors
There is good evidence that the risk of breast cancer spread to bone can be predicted both on the basis of the intrinsic genetic subtype of the primary tumour as well as the presence of recently identified bone metastasis genes.
Breast cancers can be classified into five intrinsic subtypes – luminal A, luminal B, HER2 enriched, basal-like and normal-like. Luminal-subtype tumours metastasize predominantly to bone [9, 10]. Basal-like tumours metastasize predominantly to the lymph-nodes, brain and lung, with bone being a relatively infrequent site of metastatic spread [9]. In this way, intrinsic tumour subtypes, which reflect the expression of multiple genes, can influence the probability of breast cancer spread to different target tissues.
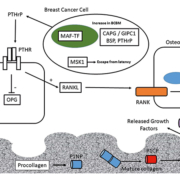
Genes that predict BCBM have been discovered using de novo unbiased genetic screening approaches – including gene copy-number analysis (CNA) – to identify regions of gene amplification specific to BCBM. In one such study, bone-homing variants of breast cancer cells were isolated by repeated intracardiac injection within immunocompromised mice and isolation of metastatic cells from bone [11]. Comparison of the parental and bone-homing cells identified a genetic region, 16q23, amplified within the bone-homing cells which encoded the gene for the musculoaponeurotic fibrosarcoma oncogene (MAF) transcription factor [11]. Further studies identified the role of MAF as a transcriptional regulator of parathyroid hormone-related protein (PTHrP) – a key regulator molecule within the vicious cycle of bone destruction within BCBM [6]. The MAF-status of primary tumours has the ability to predict the benefit of ZA treatment [12]. Patients with MAF-negative tumours have increased disease-free survival upon ZA treatment compared to control patients; however, the beneficial effects of ZA treatment are not observed in patients with MAF-positive tumours [12].
Breast cancer cells which have metastasized to bone frequently remain dormant for many years as disseminated tumour cells (DTCs). Growth signals that are still not completely understood trigger eventual activation of these DTCs and the formation of macro-metastatic lesions. In a recent study using functional genetic screening a protein kinase [mitogen and stress-activated kinase-1 (MSK1)] has been identified, which in ER-positive breast cancer cells promotes breast cancer cell differentiation and inhibits migration to bone [13]. This suggests that the level of expression of MSK1 within ER-positive breast cancer cells could be used to stratify patients in terms of risk of developing BCBM.
Protein-expression risk factors within BCBM
Several studies have focused on altered protein expression within BCBM. Immunohistochemical measurement of the levels of cyclo-oxygenase-2 (COX2), cytokeratin-5/6 (CK5/6), C-X-C chemokine receptor-4 (CXCR4), parathyroid hormone receptor-1 (PTHR1), osteoprotogerin (OPN) and calcium-sensing receptor (CaSR) within primary patient tumours evaluated their potential as potential predictors of the subsequent development of BCBM [14]. The absence of cytoplasmic OPN in this study was observed to be an independent risk factor for the development of BCBM, whereas expression of PTHR1 was observed to be associated with BCBM; however, the association was not significant within multivariate analysis, thus PTHr1 levels are not an independent predictor of BCBM [14].
Quantitative proteomic analysis of parental MDA-MB-231 triple-negative breast cancer cells and comparison with a bone-homing variant of these cells isolated by repeated intracardiac injection within immunocompromised mice, identified two proteins as predictive of development of BCBM: PDZ-domain containing protein (GIPC1) and macrophage capping-protein (CAPG) [15]. In rigorous adjusted Cox regression analyses, high expression of both CAPG and GIPC1 within primary tumours was associated with a higher risk for development of BCBM within both a training set (n=427) and a subsequent validation set (n=297) of patients selected from the large randomized AZURE trial of adjuvant ZA (AZURE-ISRCTN79831382) [15]. GAPGhigh/GIPC1high status was not associated with development of bone metastasis following ZA treatment suggesting that these two markers are also predictive of treatment benefit.
Bone morphogenetic protein-7 (BMP7) is a cytokine which can elicit diverse signalling outcomes within breast cancer cells, including altering the rates of cell migration, invasion and apoptosis, as well as its role in bone formation [16]. In a study of the level of expression of BMP7 within breast cancer primary tumours, high expression of BMP7 correlated with a reduced time to development of BCBM within invasive ductal carcinomas [17]. In this study BMP7 levels did not correlate with time to BCBM within invasive lobular carcinoma [17].
Components of the bone extracellular matrix are potential markers for BCBM risk and several proteins have been studied in this regard including bone sialoprotein (BSP), osteopontin and osteocalcin [18]. BSP is a component of the bone mineralized cell-matrix which can perform numerous functions, including integrin-binding and the regulation of angiogenesis [19]. Serum levels of BSP were observed to be higher in patients with bone-only metastasis of breast cancer compared to patients with both osseous and visceral metastases within both univariate and multivariate analysis, with a circulating BSP concentration of ≥24 ng/ml acting as a significant factor for prediction of BCBM risk [20].
Bone turnover markers to monitor development of BCBM
Bone turnover markers are products of active bone resorption and formation. Several of these markers are products of collagen metabolism including procollagen-I N-terminal extension pro-peptide (PINP) and procollagen-I C-terminal extension peptide (PICP) – markers of bone formation, as well as C-terminal type-I collagen telopeptide (CTX) and C-terminal telopeptide (ICTP) – markers of bone resorption [21]. In a study measuring the levels of P1NP, CTX and 1-CTP within 872 patient-serum samples taken at baseline in the AZURE trial of adjuvant ZA, levels of P1NP, CTX and 1-CTP were all found to be prognostic for future BCBM, but none of these markers were prognostic for non-skeletal metastasis overall survival or treatment benefit from ZA [22].
In a related study [23], Lipton et al. investigated CTX in 621 postmenopausal early breast cancer patients in a 5-year phase III trial of tamoxifen +/− octreotide. Higher pre-treatment CTX was associated with shorter bone-only recurrence-free survival. However, there was no statistically significant association with first event in the bone plus concurrent relapse elsewhere or with first recurrence at other distant sites.
In a related study serum levels of total and bone-specific alkaline phosphatase (BSAP), CTX, ICTP, osteocalcin, N-terminal telopeptide of collagen (NTX), PINP and tartrate resistant acid phosphatase (TRACP5b; a marker of bone resorption), were measured in postmenopausal women with early stage luminal-type invasive ductal carcinoma (IDC) [24]. In this study TRACP5b levels most accurately predicted the development of BCBM, with a 3-marker panel (BSAP, PINP and TRACP5b) serving as an accurate marker panel for BCBM [24].
Conclusion
The metastatic spread of breast cancer cells to bone is a multistep process in which cancer cells must enter and survive within the circulation, and then finally leave the circulation and enter (and adapt to) the bone micro-environment. Molecular profiling of breast cancer cells at both the genetic and protein level has identified a series of molecules which play pivotal roles in this complex process. As such, differential expression of these molecules within primary patient tumour samples may be used to stratify patients with early breast cancer, in terms of BCBM risk and guiding treatment decisions. To date, the intrinsic tumour subtype has proven to be the most effective observation predicting risk of BCBM development; however, recent studies have identified new molecular components within bone metastatic breast cancer cells (including key transcription factors and proteins important in cell signalling and cell migration) that may form the basis of future tests.
Once within bone, breast cancer cells trigger alterations in the bone micro-environment that favour survival of DTCs. Later when macroscopic metastases form, the altered rates of bone formation and breakdown lead to the generation of bone metabolic products that can be measured within patients. Altered levels of these bone metabolic products predict BCBM development and can also be used to monitor treatment responses. Extracellular matrix components including BSAP, PINP, TRACP5b, CTX and 1-CTP have proven particularly useful in this regard.
Studies to date have occasionally produced conflicting results. This may reflect the use of widely differing sample sources (ranging from animal model systems to patient-derived samples), as well as variations in the patient cohorts used for different clinical studies. Despite these limitations, key molecules are becoming evident that can be measured and used to predict the risk of BCBM. Future studies using these candidate molecules in larger, multicentre clinical trials will further refine a testing panel for prediction of BCBM risk.
References
1. Cancer Research UK (CRUK). Breast cancer statistics (http: //www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/breast-cancer).
2. Scheid V, Buzdar AU, Smith TL, Hortobagyi GN. Clinical course of breast cancer patients with osseous metastasis treated with combination chemotherapy. Cancer 1986; 58(12): 2589–2593.
3. Coleman RE. Metastatic bone disease: clinical features, pathophysiology and treatment strategies. Cancer Treat Rev 2001; 27(3): 165–176.
4. Wilson C, Bell R, Hinsley S, Marshall H, Brown J, Cameron D, Dodwell D, Coleman R. Adjuvant zoledronic acid reduces fractures in breast cancer patients; an AZURE (BIG 01/04) study. Eur J Cancer 2018; 94: 70–78.
5. Lipton A, Fizazi K, Stopeck AT, Henry DH, Smith MR, Shore N, Martin M, Vadhan-Raj S, Brown JE, et al. Effect of denosumab versus zoledronic acid in preventing skeletal-related events in patients with bone metastases by baseline characteristics. Eur J Cancer 2016; 53: 75–83.
6. Guise TA, Kozlow WM, Heras-Herzig A, Padalecki SS, Yin JJ, Chirgwin JM. Molecular mechanisms of breast cancer metastases to bone. Clin Breast Cancer 2005; 5 Suppl(2): S46–53.
7. Stopeck AT, Fizazi K, Body JJ, Brown JE, Carducci M, Diel I, Fujiwara Y, Martín M, Paterson A, et al. Safety of long-term denosumab therapy: results from the open label extension phase of two phase 3 studies in patients with metastatic breast and prostate cancer. Support Care Cancer 2016; 24(1): 447–455.
8. Rathbone EJ, Brown JE, Marshall HC, Collinson M, Liversedge V, Murden GA, Cameron D, Bell R, Spensley S, et al. Osteonecrosis of the jaw and oral health-related quality of life after adjuvant zoledronic acid: an adjuvant zoledronic acid to reduce recurrence trial subprotocol (BIG01/04). J Clin Oncol 2013; 31(21): 2685–2691.
9. Huber KE, Carey LA, Wazer DE. Breast cancer molecular subtypes in patients with locally advanced disease: impact on prognosis, patterns of recurrence, and response to therapy. Semin Radiat Oncol 2009; 19(4): 204–210.
10. Ignatov A, Eggemann H, Burger E, Ignatov T. Patterns of breast cancer relapse in accordance to biological subtype. J Cancer Res Clin Oncol 2018; doi: 10.1007/s00432-018-2644-2.
11. Pavlovic M, Arnal-Estape A, Rojo F, Bellmunt A, Tarragona M, Guiu M, Planet E, Garcia-Albéniz X, Morales M, et al. Enhanced MAF oncogene expression and breast cancer bone metastasis. J Natl Cancer Inst 2015; 107(12): djv256.
12. Coleman R, Hall A, Albanell J, Hanby A, Bell R, Cameron D, Dodwell D, Marshall H, Jean-Mairet J, et al. Effect of MAF amplification on treatment outcomes with adjuvant zoledronic acid in early breast cancer: a secondary analysis of the international, open-label, randomised, controlled, phase 3 AZURE (BIG 01/04) trial. Lancet Oncol 2017; 18(11): 1543–1552.
13. Gawrzak S, Rinaldi L, Gregorio S, Arenas EJ, Salvador F, Urosevic J, Figueras-Puig C, Rojo F, Del Barco Barrantes I, et al. MSK1 regulates luminal cell differentiation and metastatic dormancy in ER(+) breast cancer. Nat Cell Biol 2018; 20(2): 211–221.
14. Winczura P, Sosinska-Mielcarek K, Duchnowska R, Badzio A, Lakomy J, Majewska H, Pęksa R, Pieczyńska B, Radecka B, et al. Immunohistochemical Predictors of Bone Metastases in Breast Cancer Patients. Pathol Oncol Res 2015; 21(4): 1229–1236.
15. Westbrook JA, Cairns DA, Peng J, Speirs V, Hanby AM, Holen I, et al. CAPG and GIPC1: breast cancer biomarkers for bone metastasis development and treatment. J Natl Cancer Inst 2016; 108(4): doi: 10.1093/jnci/djv360.
16. Alarmo EL, Parssinen J, Ketolainen JM, Savinainen K, Karhu R, Kallioniemi A. BMP7 influences proliferation, migration, and invasion of breast cancer cells. Cancer Lett 2009; 275(1): 35–43.
17. Alarmo EL, Korhonen T, Kuukasjarvi T, Huhtala H, Holli K, Kallioniemi A. Bone morphogenetic protein 7 expression associates with bone metastasis in breast carcinomas. Ann Oncol 2008; 19(2): 308–314.
18. Bahrami A, Hassanian SM, Khazaei M, Hasanzadeh M, Shahidsales S, Maftouh M, Ferns GA, Avan A. The therapeutic potential of targeting tumor microenvironment in breast cancer: rational strategies and recent progress. J Cell Biochem 2018; 119(1): 111–122.
19. Bouleftour W, Granito RN, Vanden-Bossche A, Sabido O, Roche B, Thomas M, Linossier MT, Aubin JE, Lafage-Proust MH, et al. Bone shaft revascularization after marrow ablation is dramatically accelerated in BSP-/- mice, along with faster hematopoietic recolonization. J Cell Physiol 2017; 232(9): 2528–2537.
20. Bellahcene A, Kroll M, Liebens F, Castronovo V. Bone sialoprotein expression in primary human breast cancer is associated with bone metastases development. J Bone Miner Res 1996; 11(5): 665–670.
21. Glendenning P, Chubb SAP, Vasikaran S. Clinical utility of bone turnover markers in the management of common metabolic bone diseases in adults. Clin Chim Acta 2018; 481: 161–170.
22. Brown J, Rathbone E, Hinsley S, Gregory W, Gossiel F, Marshall H, et al. Associations between serum bone biomarkers in early breast cancer and development of bone metastasis: results from the AZURE (BIG01/04) trial. J Natl Cancer Inst 2018; doi: 10.1093/jnci/djx280.
23. Lipton A, Chapman JA, Demers L, Shepherd LE, Han L, Wilson CF, Pritchard KI, Leitzel KE, Ali SM, Pollak M. Elevated bone turnover predicts for bone metastasis in postmenopausal breast cancer: results of NCIC CTG MA.14. J Clin Oncol 2011; 29(27): 3605–3610.
24. Lumachi F, Basso SM, Camozzi V, Tozzoli R, Spaziante R, Ermani M. Bone turnover markers in women with early stage breast cancer who developed bone metastases. A prospective study with multivariate logistic regression analysis of accuracy. Clin Chim Acta 2016; 460: 227–230.
The authors
Steven L. Wood MA, PhD; Prof. Janet E. Brown* BMedSci, MB BS, MSc, MD, FRCP
Academic Unit of Clinical Oncology, Department of Oncology and Metabolism,
University of Sheffield, UK
*Corresponding author
E-mail: j.e.brown@sheffield.ac.uk